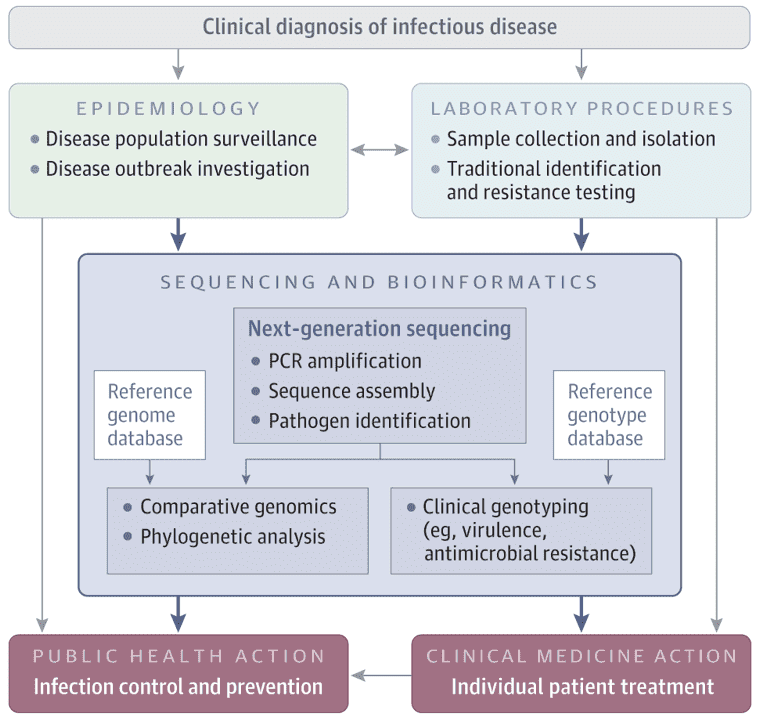
Background
Campylobacter jejuni causes acute gastroenteritis, typically from undercooked poultry or contaminated water. Severe cases might require antibiotics. AMR, driven by gene mutations and horizontal gene transfer, is a rising concern. Whole-genome sequencing (WGS) accurately identifies AMR genes and mechanisms, surpassing traditional methods. This study uses WGS to evaluate AMR and genetic diversity in C. jejuni from multiple sources.
Materials & Methods
Sample preparation:
-
Campylobacter jejuni
- DNA extraction
Data Analysis:
-
Quality control
- Assembly
- Annotation
- Phylogenomic analysis
- Resistance gene screening
Results
The genomic analysis of C. jejuni strains revealed genome sizes ranging from 1.6 to 1.86 Mb with an average G+C content of 30.32%. The pan-genome analysis identified 4204 genes, including 1215 core and 2989 accessory genes. Three clusters of C. jejuni strains were identified based on cgMLST, showing varying resistance profiles. Notably, ST-5 was predominant. WGS identified 26 AMR genes, with specific point mutations associated with resistance. Virulence genes related to motility, adherence, and stress response were found across isolates, with significant differences in gene profiles among strains from different sources.
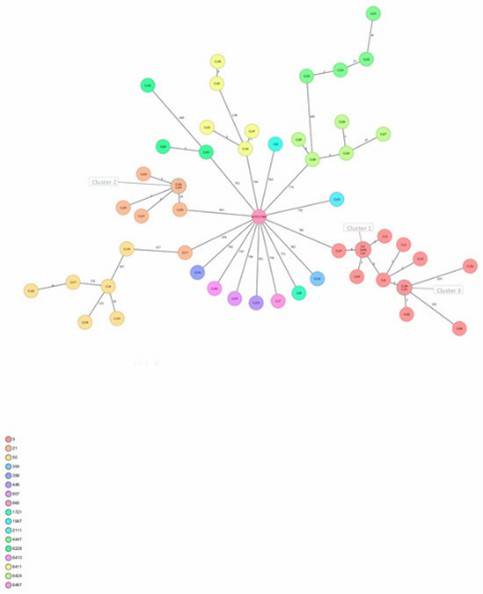 Figure 1. Minimum spanning tree of C. jejuni isolates generated.
Figure 1. Minimum spanning tree of C. jejuni isolates generated.
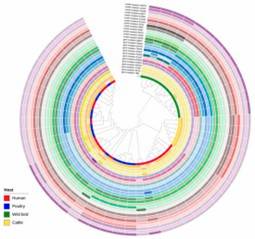 Figure 2. Interactive tree of life (iTOL) of 53 C. jejuni isolates based on the presence and absence of gene point mutations and sequence cluster classification.
Figure 2. Interactive tree of life (iTOL) of 53 C. jejuni isolates based on the presence and absence of gene point mutations and sequence cluster classification.
Conclusions
Combining WGS and cgMLST techniques revealed the spread of antimicrobial-resistant C. jejuni clones and potential transmission routes between humans and animals. The analysis showed that gene mutations linked to resistance are accumulating, indicating long-term selective pressure and evolving resistance mechanisms.


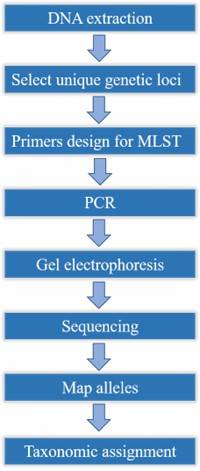
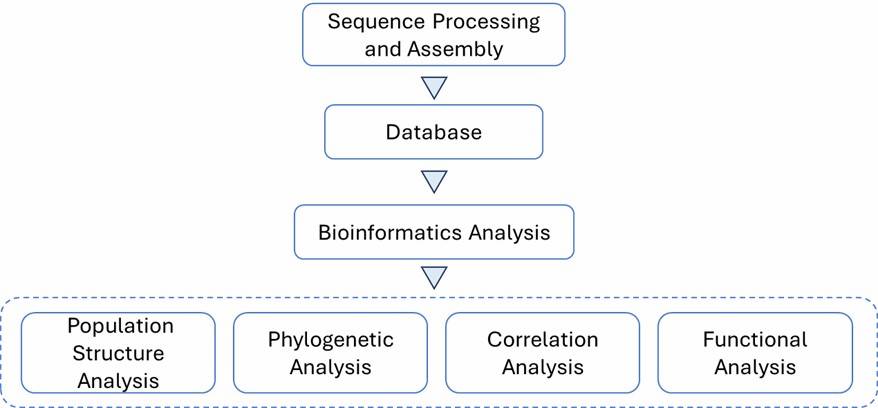
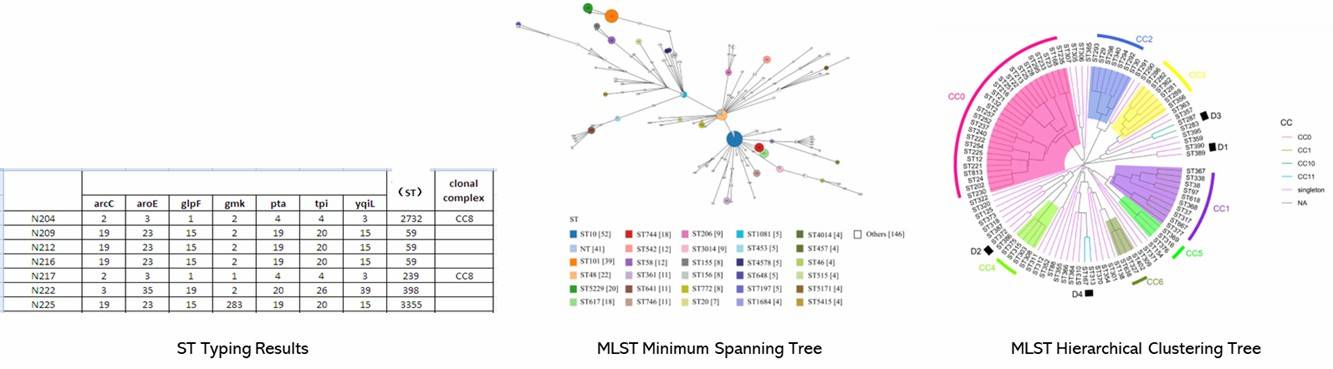

 Figure 1. Minimum spanning tree of C. jejuni isolates generated.
Figure 1. Minimum spanning tree of C. jejuni isolates generated. Figure 2. Interactive tree of life (iTOL) of 53 C. jejuni isolates based on the presence and absence of gene point mutations and sequence cluster classification.
Figure 2. Interactive tree of life (iTOL) of 53 C. jejuni isolates based on the presence and absence of gene point mutations and sequence cluster classification.