Mycobacterium tuberculosis ARGs Analysis
Service Overview
Today, antibiotic resistance genes have become an important issue of global public health concern and the biggest challenge facing infectious diseases today. Tuberculosis (TB) is a serious public health problem worldwide, and multidrug-resistant Mycobacterium tuberculosis and extensively drug-resistant Mycobacterium tuberculosis are one of the problems that need to be solved urgently. Therefore, rapid and accurate detection of drug-resistant Mycobacterium tuberculosis (MTB) in specimens is crucial to effectively control the tuberculosis epidemic. CD Genomics provides DNA microarray-based detection of antibiotic resistance genes (ARGs) in Mycobacterium tuberculosis.
Our Advantages of Mycobacterium tuberculosis ARGs Analysis:
- Eliminates the need for bacterial culture and greatly reduces the time required for antibiotic resistance gene testing.
- Provides a basis for laboratory studies of Mycobacterium tuberculosis and reduces the spread of drug-resistant strains.
- Helps to study the epidemiological distribution of MTB resistance and provides a reference basis for the prevention and treatment of Mycobacterium tuberculosis.
- The use of gene chip technology to detect antibiotic resistance gene mutations avoids false negatives, and the results are accurate and reliable.

Tell Us About Your Project
We are dedicated to providing outstanding customer service and being reachable at all times.
 Request a Quote
Request a Quote
What We Offer
Our DNA microarray-based platform can rapidly and simultaneously detect multiple resistance-related genes and identify various mutation sites associated with drug resistance. The study found that Mycobacterium tuberculosis was primarily resistant to antibiotics such as RIF-class agents, isonicotinic acid derivatives, and aminocyclitol-based compounds. The services we offer include but are not limited to the following:
- Rifamycin resistance gene detection: RIF-class agents—including RFP, RFT, and RFB—are key first-line anti-TB drugs. Our detection service targets mutations in rpoA, rpoB, and rpoC.
- Isonicotinic acid derivative resistance gene detection: Our service identifies deletions or mutations in katG, inhA, and ethA, which are associated with resistance to this class of agents.
- Arabinofuranosyl transferase-targeting drug resistance gene detection: Our platform detects overexpression or mutation in embA, embB, and embC, which are linked to resistance against ethambutol-type compounds.
- Pyrazinecarboxamide derivative resistance gene detection: We detect mutations in pncA, rpsA, and panD, genes related to resistance against PZA-class agents.
- Aminocyclitol-based agent resistance gene detection: Mutations in rpsL and rrs are assessed, especially at codons 43 and 88 of RpsL, and nucleotide positions 905 and 513 of rrs.
- Quinolone resistance gene detection: Our service targets mutations in the gyrA (A subunit) and gyrB (B subunit) genes.
- Second-line injectable agent resistance gene detection: This includes resistance to AG-class agents such as kanamycin analogs, and cyclic peptide-based agents like capreomycin-type and viomycin-type compounds. Mutations in rrs and tlyA are key markers.
- Macrolide resistance gene detection: Our detection targets mutations in ermMT and Tap.
- Cyclic amino acid derivative resistance gene detection: Resistance associated with cycloserine-type agents is detected by analyzing mutations in gadA, alrA, ddlA, and ald.
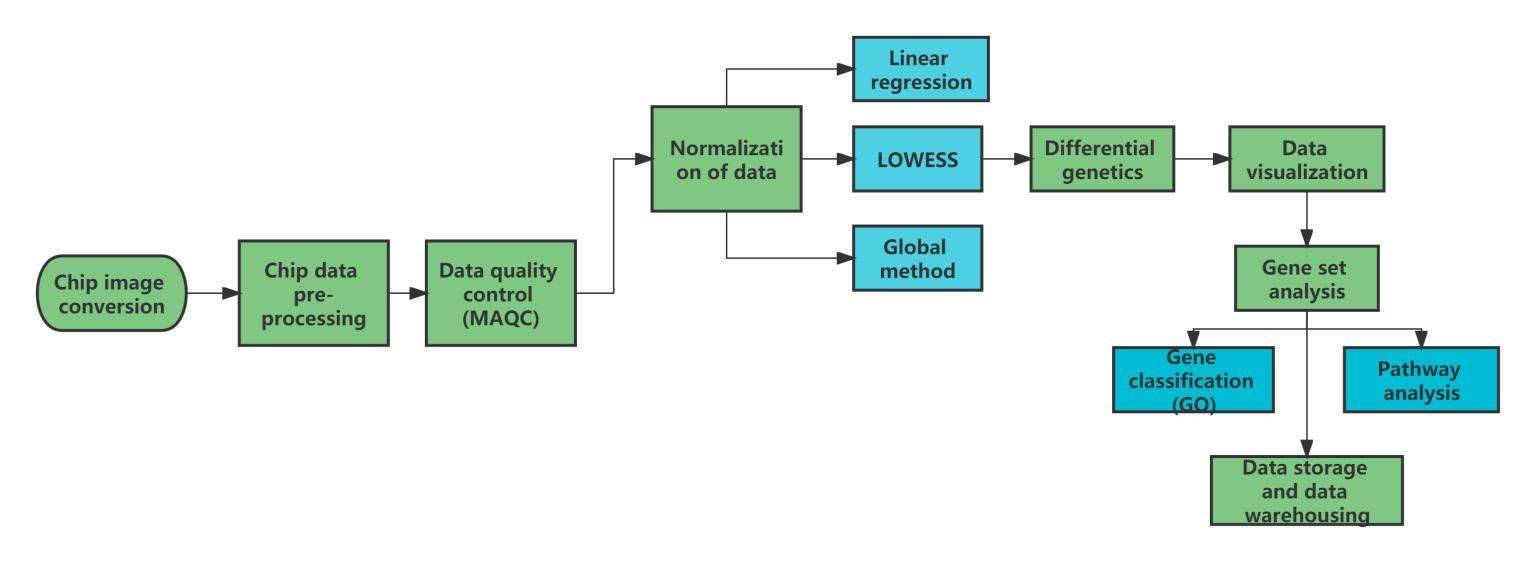
Data Analysis
Sample Requirements
| SAMPLE TYPE |
SAMPLE DETAILS |
| Sputum |
1 mL or more. For transport, use biological ice packs (2-8℃)/3 d. The container is chosen to be in a sterile airtight sputum box. |
| Alveolar lavage fluid |
2-3 mL. For transport, use biological ice packs (2-8℃)/3 d. The container is chosen to be in a sterile airtight sputum box. |
| Purulent secretions |
2-3 mL. For transport, use biological ice packs (2-8℃)/3 d. The container is chosen to be in a sterile airtight sputum box. |
| Puncture fluid |
2-3 mL. For transport, use biological ice packs (2-8℃)/3 d. The container is chosen to be in a sterile airtight sputum box. |
| Urine |
2-3 mL. For transport, use biological ice packs (2-8℃)/3 d. The container is chosen to be in a sterile airtight sputum box. |
Reference
- Tang X, Morris SL, Langone JJ, et al. Microarray and allele specific PCR detection of point mutations in Mycobacterium tuberculosis genes associated with drug resistance. J Microbiol Methods. 2005 Dec; 63(3): 318-30.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.




