In all kinds of environments in nature, there are microorganisms, and they all play a greater or lesser role in their respective "posts". Some microorganisms with special functions have attracted widespread attention due to the importance or particularity of their functions. These microorganisms with special functions are called functional microorganisms. Functional microorganisms often participate in specific geochemical cycles. The common ones are carbon cycle, Nitrogen cycle, methane cycle, sulfur cycle, etc. Common functional microorganisms include ammonia-oxidizing bacteria, ammonia-oxidizing archaea, nitrifying bacteria, nitrogen-fixing microorganisms, methanogens, sulfate-reducing bacteria, etc.
The genes that govern these functional microorganisms to perform important functions are called functional genes, such as amoA, dsrB, nxrA, nirK, mcrA, pmoA. CD Genomics has completed the detection of dozens of functional genes, and has mature detection technology and analysis experience.
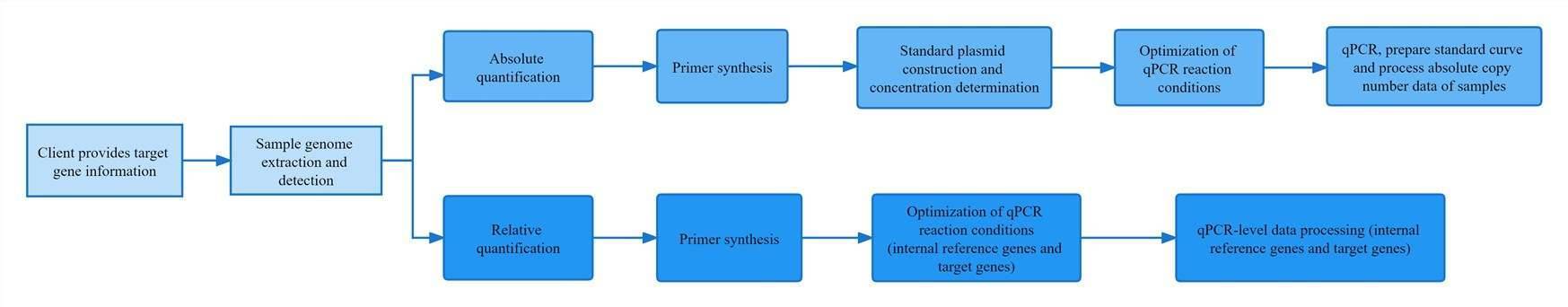
These specific functional genes are the coding genes of enzymes with a specific role, and the copy number of the gene in the sample can be determined by quantifying the gene of the enzyme by qPCR.
At present, more research is mainly on the copy number information of enzymes in each reaction in the nitrogen cycle, carbon cycle, sulfur-related, and arsenic-related enzymes.