Microbial Genomics Methods for Drug Target Discovery
Over the past years, widespread antibiotic resistance has attracted much attention since it became an important threat to modern medicine. Efforts to develop new antibiotics with novel drug targets are prioritized. Microbial genomics is a promising method to discover drug targets. Coupled with powerful bioinformatics tools, microbial genomics enables the identification, detection, selection, and validation of novel antibacterial pathways to accelerate the drug discovery process. Information on the bacterial genome sequences, DNA methylation, and gene or protein expression levels on a genome-wide scale are produced. Comparative genomic methods can identify highly conserved genes within and between bacterial species, making them attractive drug targets for the development of new antibiotics.
Accelerate Your Drug Discovery Process
According to your specific needs, we employ various sequencing platforms, including Illumina Miseq/Hiseq, Roche 454, PacBio Sequel, and Nanopore MinION systems, to gain huge and accurate microbial genomic data. We provide comprehensive information about genomic sequence and annotation of bacterial and archaeal genomes, and perform comparative genomics and evolutionary studies to find out the key genes that participate in key bacterial processes using tools such as the TIGR Comprehensive Microbial Resource (CMR) and OrthoMCL standalone software.
The Main Research Directions
- Understand how pathogens spread and cause disease.
- Identify molecular determinants of virulence and symbiosis.
- Identify new targets for therapies and antigens for vaccines.
What Can We Do?
- 1. High-throughput sequencing, such as 16S/18S/ITS sequencing, metagenomics, microbial whole genome sequencing, metatranscriptomics, et al.
- 2. Identification of promising drug targets through comparative genomics, orthology prediction, and gene essentiality prediction.
- 3. Validation of drug targets for novel antibiotics.
Note: Our service are for research use only, not for disease diagnosis or treatment.
Detectable Objects
Bacteria, fungi, archaebacteria, mycoplasma.
Detection Methods
Next-generation sequencing (NGS), long-read sequencing, PCR denaturing gradient gel analysis (PCR-DGGE), real-time qPCR.
Technical Platforms
Illumina HiSeq/MiSeq, Ion PGM, PacBio SMRT systems, Nanopore systems, PCR-DGGE, Real-time qPCR, clone library etc.
Sample Requirements
-

- DNA sample: ≥ 500 ng, OD260/280 = 1. 8 - 2. 0, concentration ≥ 10 ng/μl.
- Ensure that the DNA is not degraded. Avoid repeated freezing and thawing during sample storage and transport.
- Please use enough dry ice or ice packs during transportation.
- Nucleic acid samples should be stored at -20 °C.
Workflow
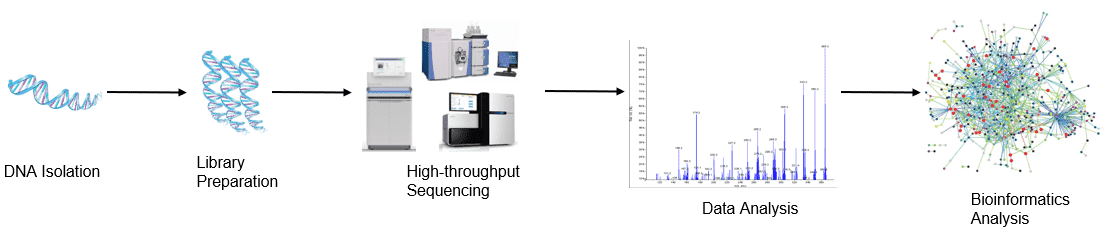 Figure 1. High-throughput sequencing analysis process.
Figure 1. High-throughput sequencing analysis process.
 Figure 2. PCR-DGGE analysis process.
Figure 2. PCR-DGGE analysis process.
Bioinformatics Analysis
| Analysis Content | Details |
| Strain diversity analysis | Understand the emergence of drug-resistant strains |
| Microbial phylogenetics | Study of epidemiology and links between genetic diversity and emergence of drug resistance |
| Comparative pathway analysis | Identification of major pathways using databases like BioCyc and KEGG |
| Comparative genomics | Identification and prioritization of essential drug targets using databases like the Database of Essential Genes and the database of Online Gene Essentiality |
References
- Qingliang Li and Luhua Lai, rediction of potential drug targets based on simple sequence properties. BMC Bioinformatics, 2007, 8:353
- Bruccoleri, R.E. et al. (1998) Concordance analysis of microbial genomes. Nucleic Acids Res. 26, 4482–4486
- Vaishali P. Waman, Sundeep Chaitanya Vedithi, Sherine E. Thomas,Bridget P. Bannerman, Asma Munir, Marcin J. Skwark, Sony Malhotra & Tom L. Blundell (2019) Mycobacterial genomics and structural bioinformatics: opportunities and challenges in drug discovery, Emerging Microbes & Infections, 8:1, 109-118