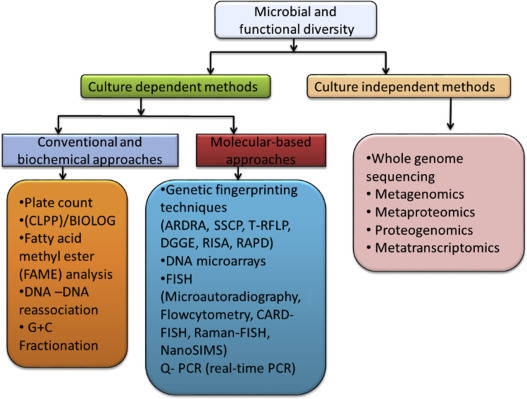
Backgrounds
Recognizing the extensive negative impacts that loss of biodiversity, precipitated by climate change, can have on ecosystem functioning and the quality and quantity of its eco-services, we turn to explore a facet of biodiversity that warrants deeper understanding - soil microbial diversity. Although previous research has illuminated the way changes in climate, specifically warming, can impact aspects such as respiration feedback responses, decomposition, biomass, community composition and succession, effects on temporal scale, and complexity and stability of networks, scant data exists when it comes to in-situ, repeat empirical observation in the field over an extensive timeline. This has resulted in a considerable gap in our knowledge about the mechanism of how warming affects subterranean microbial diversity. To narrow this gap, we embarked on a seven-year, in-situ experiment, designed to unveil how climate change - encapsulated in increased warming, alterations in precipitation, and mowing - and the interaction thereof, impact plant prairie soil bacteria, fungi and protozoa community diversity and, inherently, ecosystem functionality.
Methods
soil microbial biomass
- Soil phospholipid fatty acids (PLFA)
DNA extraction
- Sodium dodecyl sulfate (SDS)-based cell lysis
Data analyses
- Picante R package
- Linear mixed-effects models
- Structural equation modelling (SEM)
Results
1. Effects of warming on soil microbial diversity
Compared to rainfall regulation and mowing, warming exerts a significant primary impact on soil and plant variables by increasing the temperature and reducing soil moisture, concurrently lowering soil pH and augmenting NO3-. While mowing has a significantly negative effect on plant biomass, it positively impacts plant richness (Fig.2).
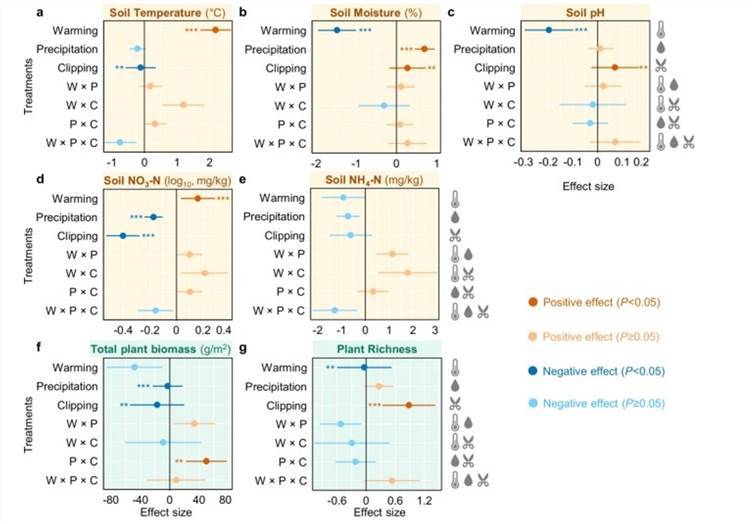 Fig. 2 Effects of experimental treatments on soil and plant variables by linear mixed-effects models (LMMs).
Fig. 2 Effects of experimental treatments on soil and plant variables by linear mixed-effects models (LMMs).
In this study, we consider two dimensions of microbial community diversity: taxonomic diversity (species richness and their relative abundance) and phylogenetic diversity. Warming has a significant negative effect on taxonomic diversity, phylogenetic diversity, and biomass across three distinct types of microbial groups (Figs. 3, 4). There were no significant feedback responses in microbial diversity to interactive effects among multiple factors, with the only exception being a positive response of fungi and protozoa to the synergistic defoliation effect accompanying warming. Therefore, the key findings suggest that warming has a dominant effect on soil bacteria, fungi, and protozoan diversity. The likely reason is that under warming conditions, changes in microbial diversity are primarily driven by alteration in the soil microenvironment and geochemistry aspects (such as soil pH, soil moisture, and soil temperature), which are significantly influenced by warming.
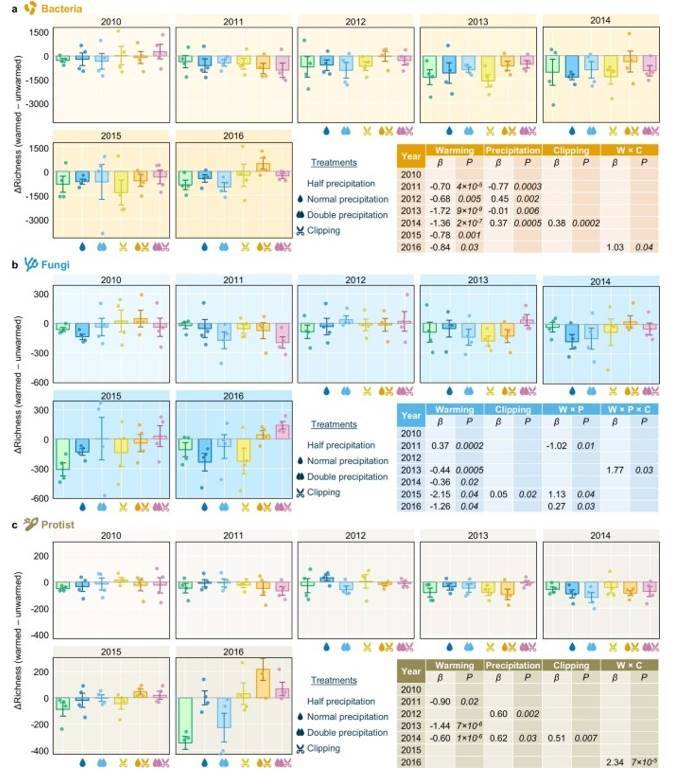 Fig. 3 Yearly differences of bacterial (a), fungal (b), and protistan (c) richness between warmed and unwarmed samples.
Fig. 3 Yearly differences of bacterial (a), fungal (b), and protistan (c) richness between warmed and unwarmed samples.
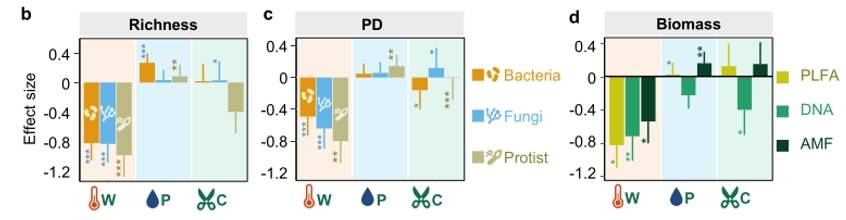 Figure 4. Effects of Different treatments on soil bacterial, fungal, and protist diversity indices, lineage diversity, and biomass
Figure 4. Effects of Different treatments on soil bacterial, fungal, and protist diversity indices, lineage diversity, and biomass
Warming exert differential effects on the microbial diversity of various lineages (Fig. 5). Within bacteria, an increase in temperature elicits a noticeable reduction in both the abundance and relative abundance of most bacterial phyla. Only the abundance and relative abundance of Actinobacteria, as well as the relative abundance of Firmicutes and Bacillus, manifest a significant positive feedback. A possible explanation for this may be their ability to form spores, which helps them withstand desiccation stress and therefore becomes more adaptive to relatively dry soils. In regard to fungi and protozoa, warming also markedly imposes negative impact on their taxonomic diversity and phylogenetic diversity, except for an evident positive feedback on both abundance and phylogenetic diversity of the Conosa in protozoans.
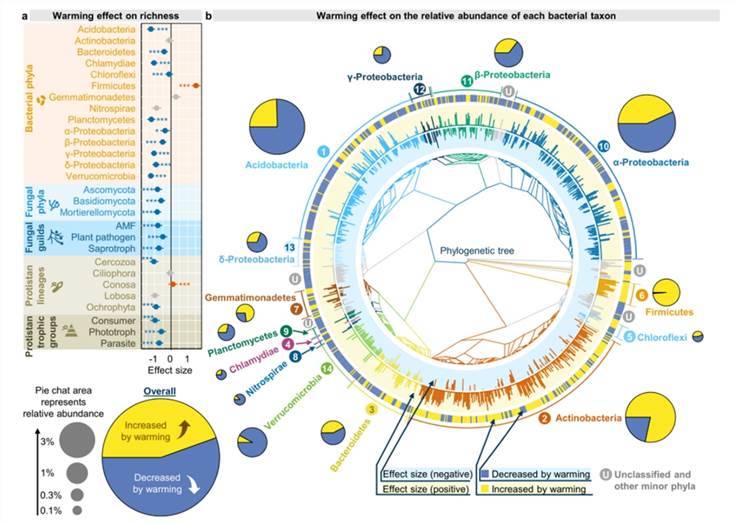 Fig. 5. Effects of experimental warming on different microbial taxa.
Fig. 5. Effects of experimental warming on different microbial taxa.
2. The Mechanisms of Microbial Diversity Reduction
At the inception of this study, we hypothesized that a warming climate would diminish microbial diversity by altering environmental conditions and biotic interactions. Our findings, drawing on a 2021 study by our research team, demonstrated that sustained warming heightened the complexity and convolutedness of the microbial species network. This further elucidates that global warming fosters competitive relations amongst microorganisms, eventually leading to the extinction of less competitive species and subsequent reduction in microbial diversity.
Warming acts as a decisive filtering factor in our research, positively selecting for microorganisms related to spore formation and negatively selecting for those unrelated to this process. This aligns synergistically with previous research findings. Consequently, to unveil the driving mechanisms behind the warming-induced decline of microbial diversity in this study, we employed Structural Equation Modeling to analyze the direct and indirect effects of environmental drivers on microbial diversity, as illustrated in Figure 6.
This model accounted for over half of the interpretation in microbial diversity, therefore leading to the conclusion that the environmental filtering effect, by influencing microbial activities and interactions, serves as the primary driving factor in the decline of microbial diversity.
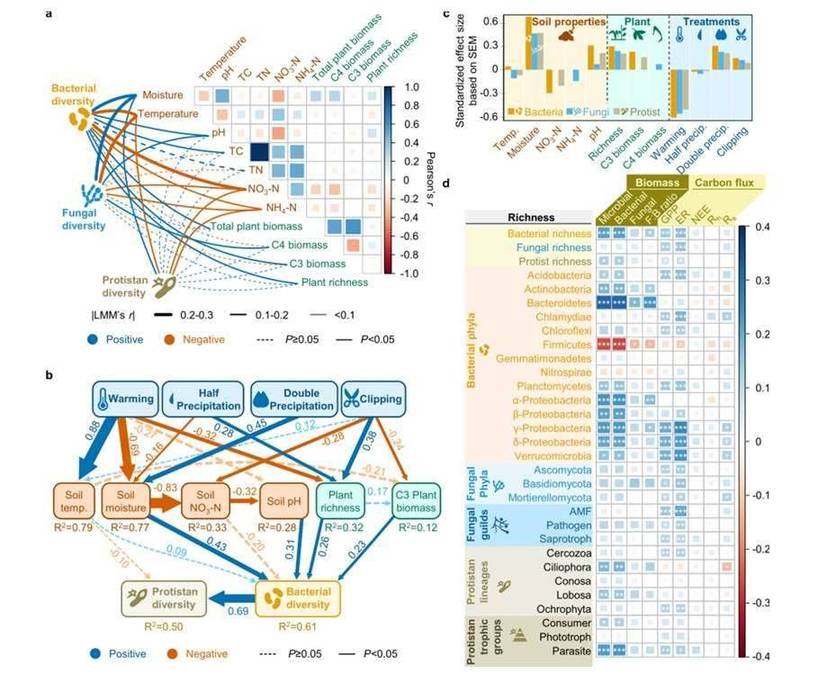 Fig.6. Environmental drivers of microbial diversity.
Fig.6. Environmental drivers of microbial diversity.
3. The Relationship Between Microbial Diversity and Ecosystem Functioning"
Beyond the domain of solely documenting warming-reduced microbial diversity, the current study bridges ecological functional process, coaxing an understanding of how alterations in microbial diversity due to climate change could, in turn, influence the ecological functional process. Observations indicate that warming downgrades various ecological functions such as total microbial biomass, bacterial biomass, primary productivity, and ecosystem respiration (refer Figure 7). Additionally, a significant positive correlation was found between microbial diversity and the ecological functional process (as depicted in Figure 6).
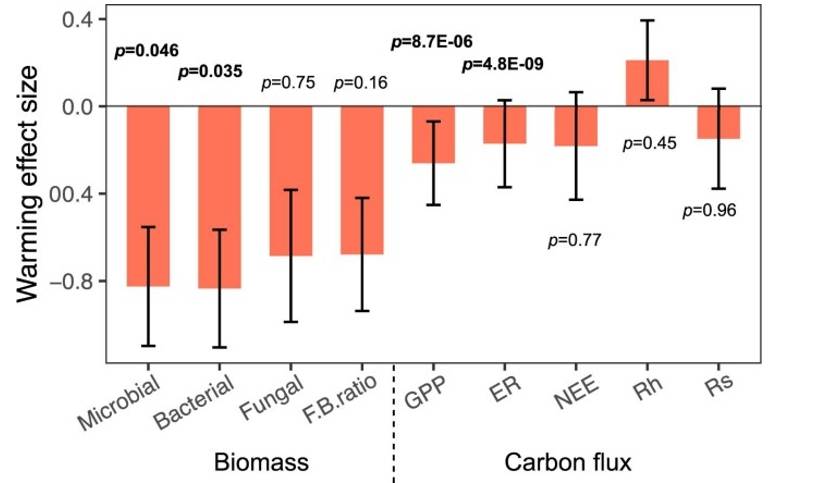 Fig. 7 Effects of experimental warming on different ecosystem functions.
Fig. 7 Effects of experimental warming on different ecosystem functions.
Conclusion
The impacts of global warming, acting as a decisive filtering factor, instigates aridity stress. Subsequently, it manipulates microbial activity and interplay, instigating a decline in microbial diversity and concomitantly rendering the associated ecosystem functions more vulnerable. Notably, the effects of increased temperature vary immensely across different microbial taxa. For instance, beneficial fungi within an ecosystem, Arbuscular Mycorrhizal Fungi (AMF), tend to decline due to warming, hence compromising the associated ecosystem functions under a future warming climate. Furthermore, it appears that the soil moisture reduction largely mediates the microbial diversity response to warming. Thus, in global drylands, encompassing arid, semi-arid, and dry sub-humid ecosystems, the impact of global warming on microbial diversity loss might be more severe. Consequently, investigating further whether the decline in microbial diversity due to warming and its corresponding driving mechanisms apply to other ecosystems is warranted.



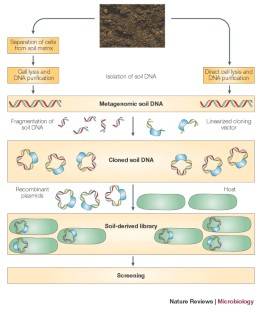
 Figure 1. High-throughput sequencing analysis process.
Figure 1. High-throughput sequencing analysis process. Figure 2. PCR-DGGE analysis process.
Figure 2. PCR-DGGE analysis process.

 Fig. 2 Effects of experimental treatments on soil and plant variables by linear mixed-effects models (LMMs).
Fig. 2 Effects of experimental treatments on soil and plant variables by linear mixed-effects models (LMMs). Fig. 3 Yearly differences of bacterial (a), fungal (b), and protistan (c) richness between warmed and unwarmed samples.
Fig. 3 Yearly differences of bacterial (a), fungal (b), and protistan (c) richness between warmed and unwarmed samples. Figure 4. Effects of Different treatments on soil bacterial, fungal, and protist diversity indices, lineage diversity, and biomass
Figure 4. Effects of Different treatments on soil bacterial, fungal, and protist diversity indices, lineage diversity, and biomass Fig. 5. Effects of experimental warming on different microbial taxa.
Fig. 5. Effects of experimental warming on different microbial taxa. Fig.6. Environmental drivers of microbial diversity.
Fig.6. Environmental drivers of microbial diversity. Fig. 7 Effects of experimental warming on different ecosystem functions.
Fig. 7 Effects of experimental warming on different ecosystem functions.