In the landscape of progressing scientific inquiry, there arises a heightened significance in comprehending the mechanisms and evolutionary dynamics of biological processes. This pertains to a holistic and intricate examination of the interplay among microbes, hosts, and the environment. Consequently, the adoption of multi-omics methodologies emerges as an invaluable tool, facilitating a thorough dissection of the logical intricacies interwoven within datasets. This approach expedites the identification of pivotal mechanisms underpinning phenotypic distinctions across diverse sample cohorts, concurrently unveiling associations with microorganisms and metabolites.
Within the realm of environmental microbial community analysis, investigations into inter-species associations stand as pivotal endeavors, showcasing not only significant importance but also a high degree of cost-effectiveness. This integrative approach facilitates a comprehensive exploration of microbial mechanisms, delving into the intricacies from both bacterial and fungal vantage points. The multidimensional facets of this research encompass analyses of species diversity, structural considerations, and the influence of environmental factors, among others. A granular examination of the interplay between bacteria and fungi unveils nuanced species-network relationships, particularly in contexts like substrate competition. This exposition how to use 16S + ITS correlation analysis to study species-species correlations of environmental microbial community analysis.
Research Approach of Species Correlation: 16S+ITS
Regarding the methodological approach to investigating species correlation, the utilization of both 16S and ITS sequencing analyses across all samples broadens the horizons of the research scope.
CD Genomics has developed a robust and verified procedure for 16S + ITS correlation analysis.
Our technicians have a wealth of experience in meta-project research; non-process analysis to ensure the practicality and accuracy of data results; and personalized information analysis program development to ensure that customer needs are met.
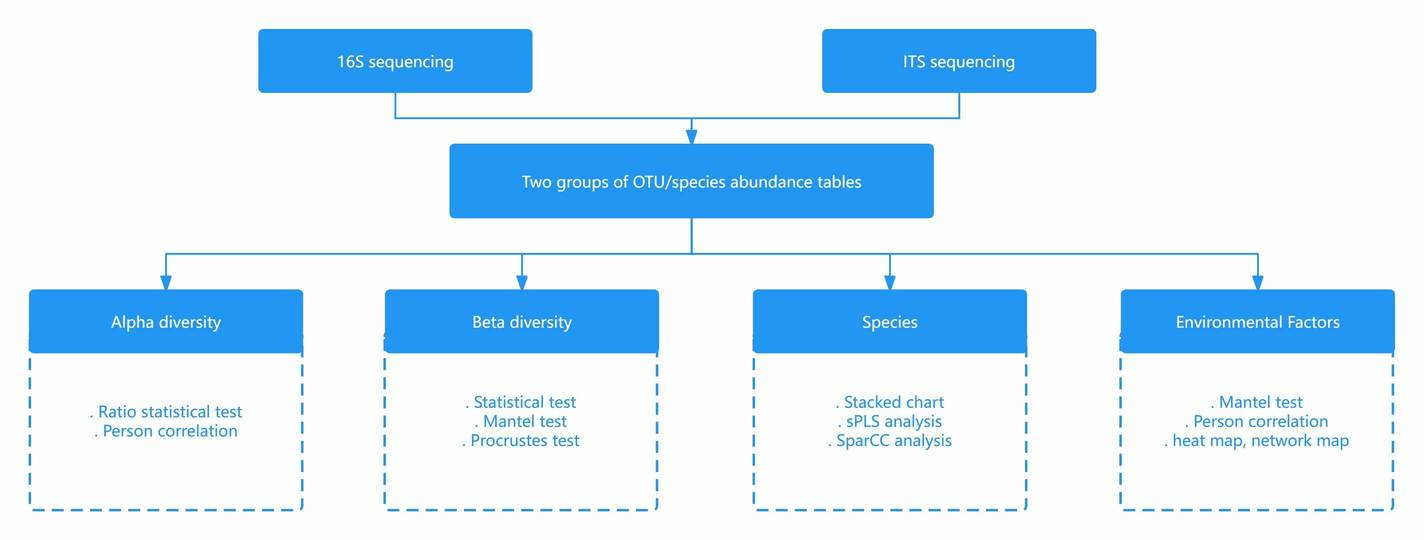 CD Genomics' research approach on correlation analysis of 16S and ITS sequencing
CD Genomics' research approach on correlation analysis of 16S and ITS sequencing
Common Techniques for Species-Species Correlation Studies
16S Amplicon Sequencing: This approach is employed for the detection of either the complete 16S sequence or its highly variable regions. The primary aim is to extract structural information on bacterial populations from environmental microbes and extrapolate predictions regarding gene functionality. Common applications involve the analysis of bacterial composition, variance, and community structure within environmental microbial communities. To learn more about 16S Amplicon Sequencing, refer to the articles "16S rRNA Sequencing VS Shotgun Sequencing for Microbial Research" and "MicroSEQ Identification by 16S/D2 Region Sequencing: Introduction, Protocol, and Applications"
ITS Amplicon Sequencing: Utilized for detecting the highly variable region within ITS, this technique aims to elucidate the structural aspects of the fungal population within environmental microbes. It finds application in the analysis of fungal composition, variations, and community structure within environmental microbial communities. To understand what ITS Amplicon Sequencing is, refer to the article "ITS Sequencing in Microbiological Research: Introduction, Bioinformatics, and Applications"
Data Analysis of Species Associations
16s data analysis
1) This method captures the comprehensive bacterial distribution in environmental microbial communities and identifies bacterial markers distinguishing various comparison groups.
2) It enables the exploration of dynamic network interactions among bacteria, facilitating a thorough understanding of the pivotal role of key bacteria and dynamic network changes within the entire bacterial community.
3) The analysis establishes connections between key bacteria and environmental factors, providing further insights into bacteria closely linked to these environmental factors.
ITS data analysis
1) Acquire the overall distribution of fungi in environmental microbial communities, along with fungal differential markers between distinct comparison groups.
2) Elucidate the dynamic network interactions among fungi, providing a comprehensive understanding of the pivotal role of key fungi within the entire fungal community and the dynamic changes in network patterns.
3) Establish the relationships between key fungi and environmental factors, further analyzing key fungi closely associated with specific environmental factors.
Correlation analysis of 16S and ITS sequencing
1) Through a comprehensive comparison of bacteria and fungi, we can thoroughly investigate the variations and commonalities in their responses to group differences, considering aspects of species diversity and community structure.
2) Additionally, exploring the networks of interaction among bacteria, fungi, and their interrelations allows for a nuanced understanding of the dynamic network changes in these crucial species within the entire microbial community. The mutual influences exerted by these key bacteria and fungi within diverse dynamic interaction models offer substantial insights for ecological investigations of the complete environmental community.
3) Moreover, conducting correlation analyses between key bacterial and fungal species and phenotypic information related to the environment can unveil their intricate relationships with environmental factors. This comprehension facilitates a comprehensive analysis of the dynamic changes observed in distinct bacteria or fungi under comparable or divergent environmental conditions. Consequently, it aids in pinpointing the primary environmental influencers and enhances our understanding of how these environmental factors impact the occupation of ecological niches and the evolution of species.
Conclusion
The investigation of inter-species relationships in environmental microbial communities should ideally revolve around three primary components:
1) Initially, the differential bacteria can be identified from the results of 16S analysis, providing insights into the network interactions between the bacterial species. By associating environmental factors with the data, primary environmental variables influencing the bacterial community can be located.
2) Subsequently, similar methods can be used with the outcomes of ITS analysis to find differential fungi, thus teasing out the network interactions between fungal species. Identifying key environmental variables that play a significant role in fungal communities also forms part of this stage.
3) Finally, an integrated comparison of the bacterial and fungal communities should be undertaken for corresponding analysis points. This comparison will explore the interplay between bacteria and fungi, interrogating existing inter-species networks and the underlying biological implications.