ITS Sequencing in Microbiological Research: Introduction, Bioinformatics, and Applications
Inquiry >Introduction to ITS Sequencing
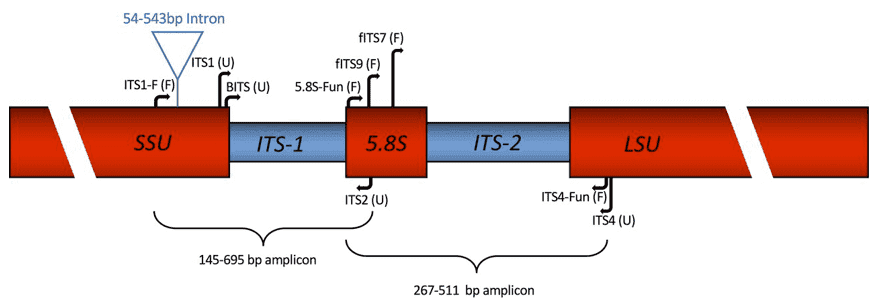
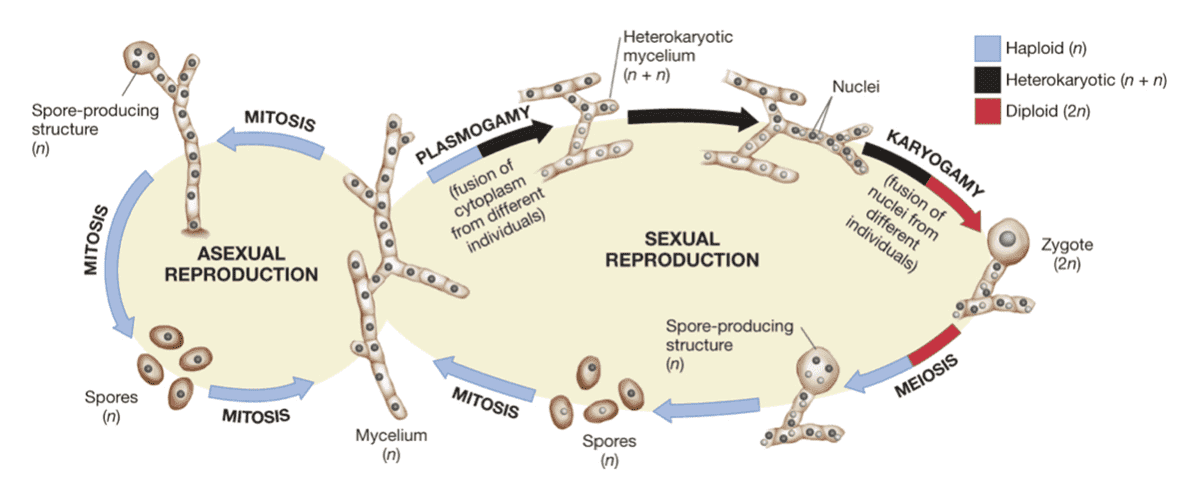
Between greatly preserved nuclear ribosomal DNA (rDNA) genes is the Internally Transcribed Spacer (ITS) (18S, 5.8S, 28S). It is a viable marker for species identification because it is hypervariable and distinctive among species but mildly unchanged among individuals from the same species. The distinctions between fungal genera, species, and even strains can be reflected by ITS. Furthermore, ITS sequence fragments are short (ITS1 and ITS2 are 350 and 400 bp, respectively) and comprise interspecific polymorphisms. Because the fragments are short, they are convenient to amplify, sequence, and analyze. Advanced NGS platforms (Illumina PE250/300) are ideal for identifying and quantifying known fungi, as well as discovering new ones.
Fungal ITS sequencing can detect sample diversity, allowing researchers to investigate the biological importance of the specimens. Fungal ITS sequencing has become crucial for the quick and accurate identification of fungi in a routine diagnostic microbiology laboratory as a result of the emergence of antifungal drug-resistant fungi. The ITS sequence of fungi can also be used to diagnose fungal plant diseases. Environmental conservation, agricultural production, petroleum exploration, and industrial manufacturing are just a few of the fields where this approach has been used.
Bioinformatics Analysis of ITS Sequencing
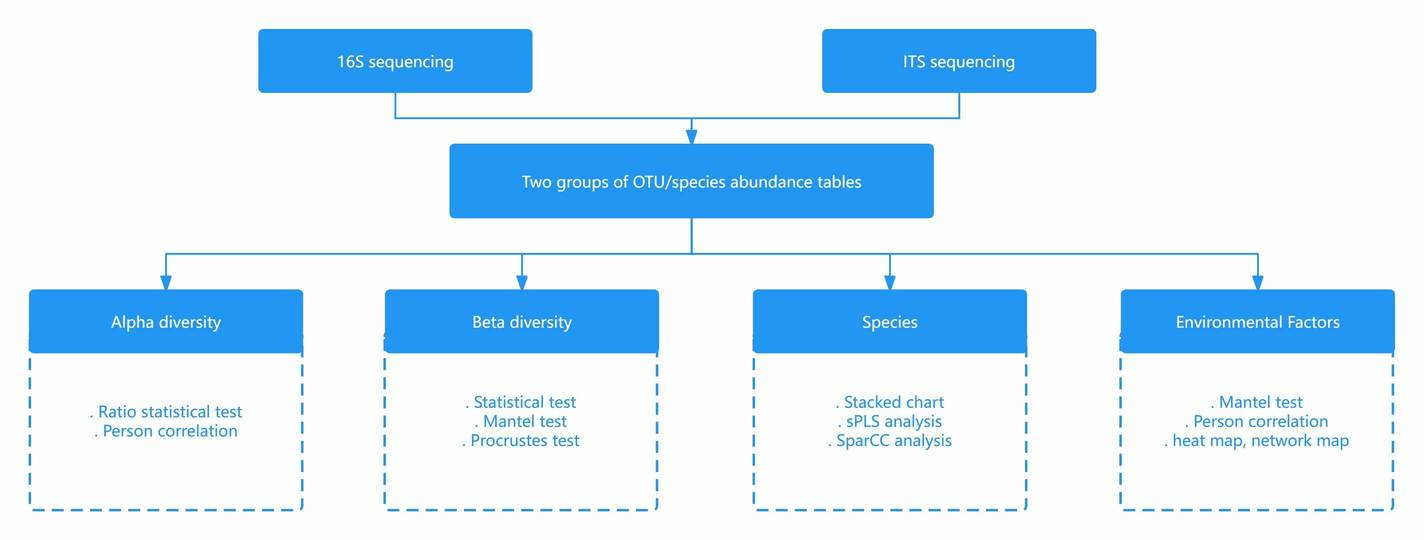
There are five main processes in the bioinformatic analysis of ITS sequencing: (1) raw data processing, (2) taxonomic assignment, (3) diversity and richness analysis, (4) comparative analysis, and (5) evolutionary analysis.
Filtering and trimming of low-quality sequences, as well as read alignment, are all part of raw data processing. The operation taxonomic unit clustering is then added to the taxonomic assignment. The rank abundance, Venn, alpha diversity, beta diversity, rarefaction curves, and heatmap are used to analyze diversity and richness. Comparative analysis, on the other hand, employs metastats, LEfSe, PCA analysis, and PCoA analysis. Finally, phylogenetic trees and (un)weighted UniFrac analysis are used in evolutionary analysis. Other custom analyses, such as CCA analysis, PICRUSt, and network analysis, are possible.
Applications of ITS Sequencing
The connection between human microorganisms and human health or illness can be studied using ITS sequencing:
- Microorganisms of the agricultural rhizosphere, plant interactions, fertilization, and the microbial community of the soil
- Study on animal intestines, animal health, and nutrition digestion.
- Sewage diagnosis, marine pollution, and petroleum degradation are all subjects of study.
- Microbial communities in extreme environments like glaciers and volcanoes.
Use in Phylogeny
Due to several advantages, sequence comparison of the eukaryotic ITS regions is widely used in taxonomy and molecular phylogeny:
- Because of its small size and the availability of highly conserved flanking sequences, it is frequently amplified.
- The high copy number of the rRNA clusters makes it easy to detect even from small amounts of DNA.
- Through unequal crossing-over and gene conversion, it undertakes fast coordinated evolution. Although high-throughput sequencing revealed frequent variants within plant species, this facilitates intra-genomic homogeneity of the repeat units.
- Even among closely related species, there is a lot of variation. This can be stated by the fact that non-coding spacer sequences are subjected to relatively low evolutionary pressure.
Mycological barcoding
The ITS area has been proposed as the universal fungal barcode sequence because it is the most widely sequenced DNA area in the molecular ecology of fungi. It has traditionally been used for molecular systematics at the species to genus level, as well as within species. Variance among individual rDNA repeats can occasionally be noted within both the ITS and IGS areas, due to its higher degree of the variance than other genic areas of rDNA. Several taxon-specific primers have been defined that enable selective amplification of fungal sequences, in addition to the universal ITS1+ITS4 primers utilized by many laboratories. Despite the fact that shotgun sequencing techniques are becoming more popular in microbial sequencing, the low biomass of fungi in clinical specimens makes ITS region amplification a research topic.
References
- Yang RH, Su JH, Shang JJ, et al. Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PloS one. 2018 Oct 25;13(10).
- Ryberg M, Kristiansson E, Sjökvist E, Nilsson RH. An outlook on the fungal internal transcribed spacer sequences in GenBank and the introduction of a web‐based tool for the exploration of fungal diversity. New Phytologist. 2009 Jan;181(2).