The importance of genital tract microbiome
The microecological environment in the reproductive tract plays an important role in women's health, reproductive function, and baby birth. A large number of normal microorganisms are present in the reproductive tract, adhering to the vaginal epithelial cells through the adhesion mechanism growing on the surface of the vaginal mucosa. Microbes in the reproductive tract interact with each other and are in a dynamic equilibrium state affecting not only the microbiome but also the vaginal environment. Studies have shown that reproductive tract microbiome can be affected by a variety of factors, including hormonal changes, pathogen invasion, antibiotic drugs, and behavior.
Accelerate research and practice in genital tract microbiome
We can provide specific information about the diversity, abundance, evolutionary relationship of reproductive tract microbes, and the interaction among microorganisms or between the microbiome and the environment using next-generation sequencing (NGS), PacBio SMRT sequencing, or Nanopore sequencing technology coupled with powerful bioinformatics analysis. In addition, we also utilize real-time qPCR to qualitatively and quantitatively study the reproductive tract microbiome.
The main research direction of genital tract microbiome
- Exploring the differences in reproductive tract microbiome between the diseased group and healthy group
- Studying the differences of reproductive tract microbiome under different environment or drug level
- Studying the microbial changes during the development of reproductive tract diseases
- Exploring the reproductive tract microbiome in the same population at different developmental stages
What can we do?
- 1. Identification and discovery of microorganisms in the reproductive tract
- 2. Quantitation of microorganisms in the reproductive tract
- 3. Evolutionary and functional analysis of microorganisms in the reproductive tract
- 4. Prediction of microbe–disease association
Note: Our services are for research use only, not for disease diagnosis and treatment.
Detectable Objects
We have successfully analyzed reproductive tract microbiome samples from human, mouse, rat, monkey, chicken, sheep, fish, horse, donkey, panda, pangolin and other animals.
Detectable Microorganisms
Bacteria (such as lactic acid bacteria), fungi, viruses, etc.
Technical Platforms
We are equipped with High-throughput sequencing, Ion PGM, PacBio SMRT systems, Nanopore systems, real-time qPCR, PCR-DGGE, and other detection platforms.
Sample Requirement

DNA sample: total DNA ≥ 300 ng, concentration ≥ 10 ng/μL, OD260/280=1.8-2.0
Make sure the DNA is not degraded and do not freeze and thaw the DNA repeatedly.
Please prepare enough ice pack or dry ice to ensure low temperature during transportation.
Workflow
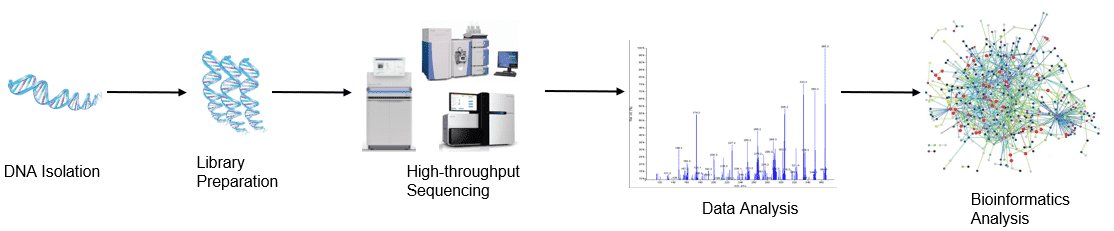 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process
Bioinformatics Analysis
| OTC Generation | OTU Clustering and Species annotation |
| Rarefraction Curve | |
| Shannon-Wiener Curve | |
| Relative Abundance | Veen Diagram |
| Heatmap | |
| Principal Components Analysis (PCA) | |
| Matastats Analysis | |
| Community Structure | Principal Coordinate Analysis (PCoA) | CCA/RDA Analysis |
| Multiple Contrast | |
| (Un)Weighted Unifrac Test | |
| Rank-Abundance Curve | |
| Structure of Microbial Colony Images | |
| Phylogenetic Tree |