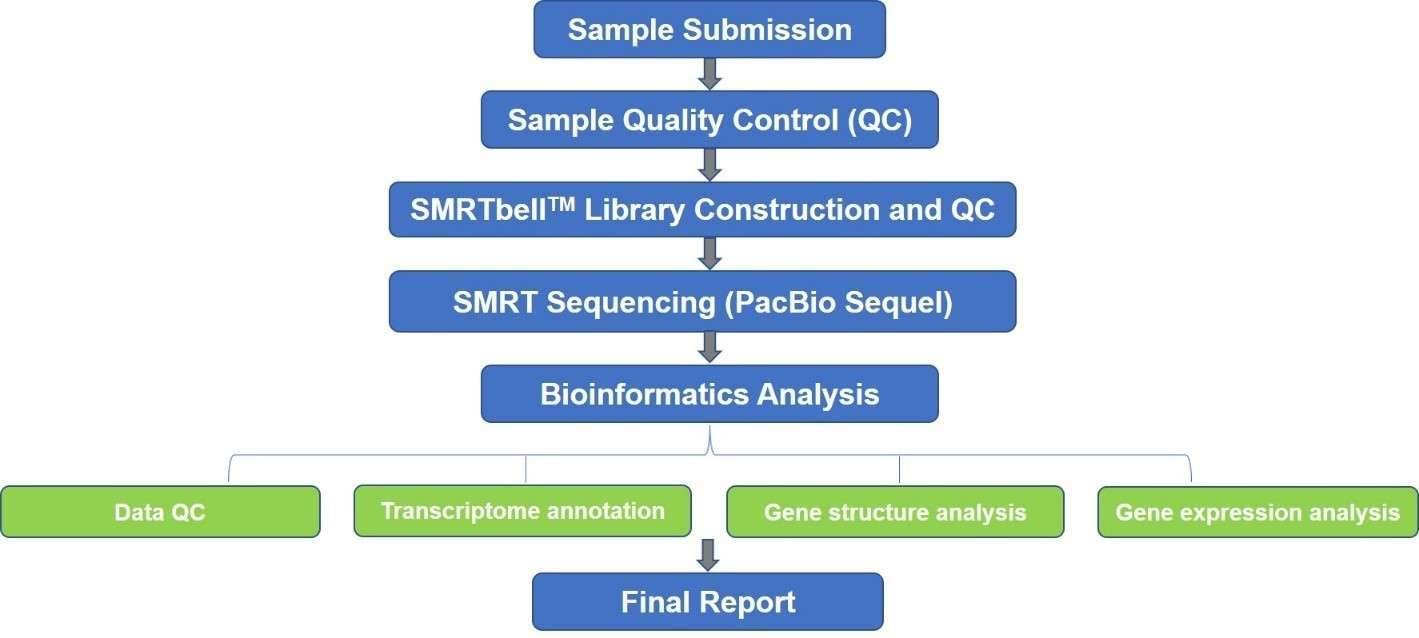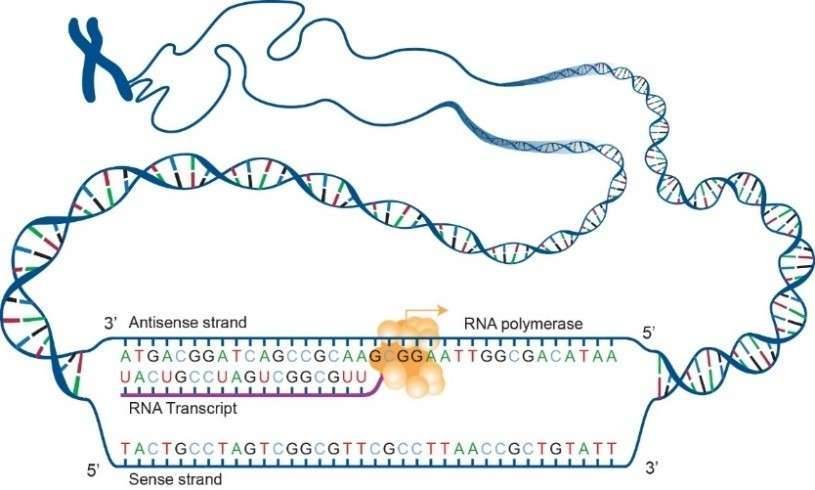
Transcriptomics enables genome-wide analysis of transcription at single-nucleotide resolution, including determination of the relative abundance of transcripts, unbiased identification of alternative splicing events and post-transcriptional RNA editing events, and detection of single nucleotide polymorphisms (SNPs). While analyzing co-post-transcriptional processing events is difficult with the short-read sequencing, SMRT isoform sequencing (Iso-Seq) is a long-read sequencing strategy that can span the full-length of transcripts. It is able to sequence the full-length isoforms without the need for assembly.
Our SMRT-based transcriptomics platform represents an easy and accurate way for various applications, like gene annotation, identification of transcript isoforms and fusion transcripts, long non-coding RNA (lncRNA) discovery, microRNA (miRNA) discovery, and prediction of potential mRNA target molecules. It also helps improve genome annotation by identifying gene structures, coding regions, and regulatory elements. Additionally, we also provide Nanopore-based microbial epigenomics service. Our SMRT-based transcriptomics can be used to address a variety of research questions, like early embryo development, phylogenetic inference, cellular differentiation, biomarker discovery, identification of drug targets, etc.