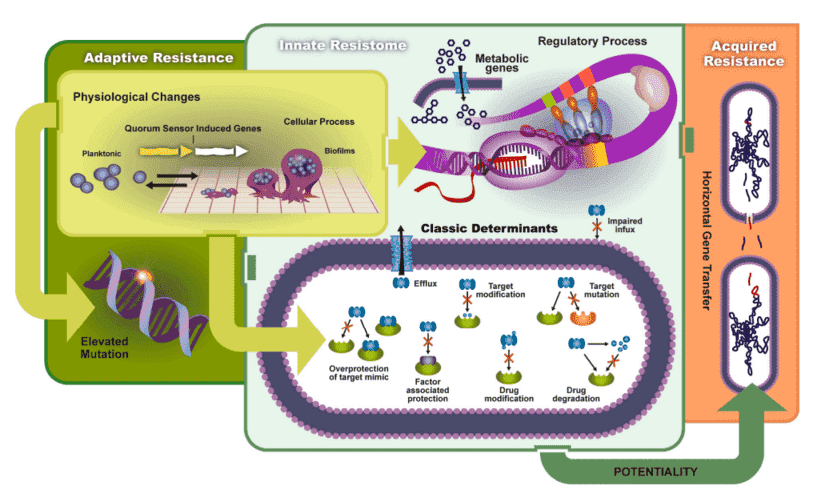
Antibiotic resistance is a major public health threat, increasing mortality and straining healthcare systems. The identification of β-lactamase genes is crucial, as these enzymes impact the effectiveness of widely used antibiotics. PCR and its derivatives are the gold standard for ARG detection, applied in over 90% of ARG studies for pre-amplification, detection, and validation, playing a key role in resistance gene research.
Our Advantages:
- Detailed Gene Targeting: Accurately identifies β-lactamase genes, including AmpC β-lactamases, for precise resistance profiling.
- Phenotypic Supplement: Enhances traditional phenotypic methods for detecting multidrug-resistant microbes.
- Efficient Surveillance: Supports large-scale surveillance and epidemiological monitoring with high-throughput PCR solutions.
- High Sensitivity: PCR-based methods provide precise detection of low-abundance ARGs in diverse microbial samples.
- Rapid Turnaround: Provides fast and efficient results, ideal for large-scale research studies.
- Comprehensive Profiling: Multiplex PCR detects up to 50 ARGs simultaneously, ensuring broad resistance gene analysis.
Introduction to Antibiotic Resistance Gene Detection Methods
The detection of antibiotic resistance genes (ARGs) is paramount for the surveillance and understanding of the propagation of antibiotic-resistant bacteria. A diverse array of methodologies exists to identify and quantify these genes, each with specific strengths and applications. This article provides an overview of the primary detection methods utilized in the field.
1. Polymerase Chain Reaction (PCR)
PCR remains a foundational technique for the amplification of targeted DNA sequences and is central to ARG detection. The principal PCR-based methods include:
- Conventional PCR: This method amplifies target ARGs, with detection typically achieved via gel electrophoresis. While effective for confirming the presence of ARGs, conventional PCR does not yield quantitative data.
- Real-Time Quantitative PCR (qPCR): Utilizing fluorescent dyes or probes, qPCR tracks the amplification process in real-time, thus providing both qualitative and quantitative data. Its high sensitivity and accuracy render it the gold standard in ARG detection.
- Multiplex PCR: This technique allows for the concurrent detection of multiple ARGs within a single reaction by employing multiple primer sets, thereby enabling comprehensive resistance profiling.
- High-Throughput Quantitative PCR (HT-qPCR): This advanced method facilitates the simultaneous analysis of numerous samples and ARGs using multiple primer pairs, optimizing efficiency and throughput in large-scale studies.
2. Gene Sequencing
Gene sequencing, encompassing methods such as Sanger sequencing and Next-Generation Sequencing (NGS), offers exhaustive identification and analysis of ARGs through the determination of DNA nucleotide sequences. NGS, in particular, is notable for its capability to detect a wide array of both known and novel ARGs. Metagenomic sequencing, a subset of NGS, further enhances this by enabling the identification of ARGs directly from environmental samples, thus providing a comprehensive overview of resistance genes present in complex microbial communities.
3. Isothermal Amplification
Isothermal amplification techniques, including Loop-Mediated Isothermal Amplification (LAMP) and Recombinase Polymerase Amplification (RPA), enable DNA amplification at a constant temperature. These methods provide rapid and straightforward alternatives to traditional PCR.
4. Fluorescence-Based Techniques
Fluorescence-based techniques involve the use of fluorescent dyes or probes for the detection and quantification of ARGs. Techniques such as Fluorescence In Situ Hybridization (FISH) and fluorescent PCR fall into this category and are noted for their sensitive detection capabilities.
How does PCR detect antibiotic resistance
Principles of PCR: PCR constitutes a pivotal molecular biology technique designed for the amplification of specific DNA sequences. This technique comprises a series of repeated thermal cycles involving denaturation, annealing, and extension phases. Within the framework of antibiotic resistance gene (ARG) detection, PCR specifically targets genes that are known to confer resistance to antibiotics. By amplifying these resistance-associated genes, PCR facilitates not only their identification but also their quantification within a given biological sample.
Multiplex PCR is a sophisticated adaptation of conventional PCR that permits the concurrent detection of multiple ARGs in a single reaction. This technique employs an array of primers, each specific to distinct ARGs, thereby enabling the simultaneous amplification of several resistance genes. The resultant PCR products can be distinguished either by their respective sizes or through the use of different fluorescent dyes in real-time PCR (RT-PCR) assays. This multiplex approach is especially advantageous for comprehensive resistance profiling and is highly suited to high-throughput screening applications.
Partial results of our PCR-based antibiotic resistance gene analysis service are shown below:
Please feel free to reach out if you have any further inquiries or require additional information.



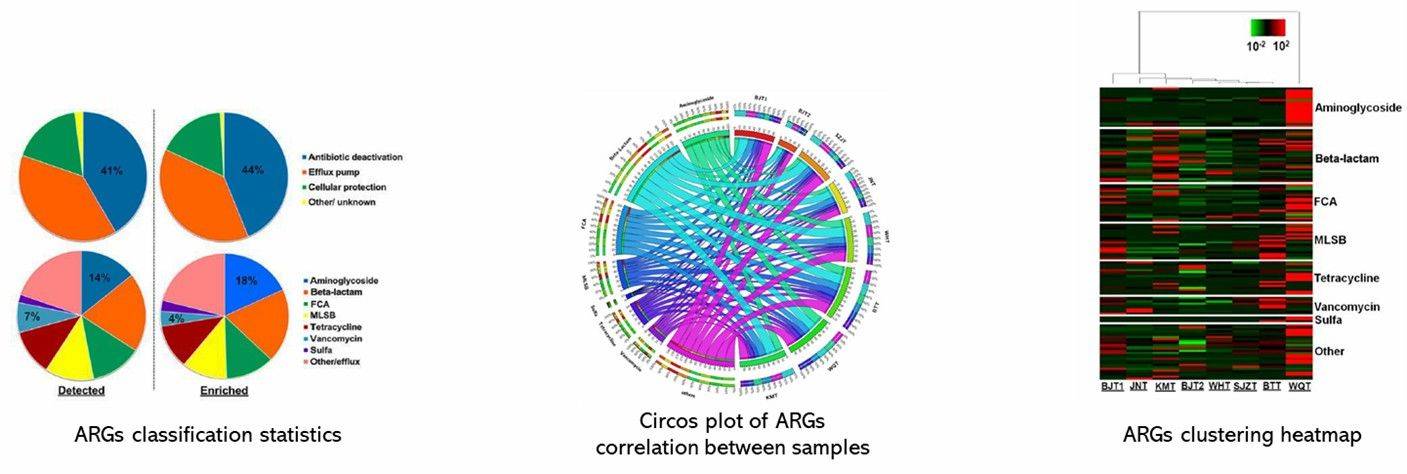

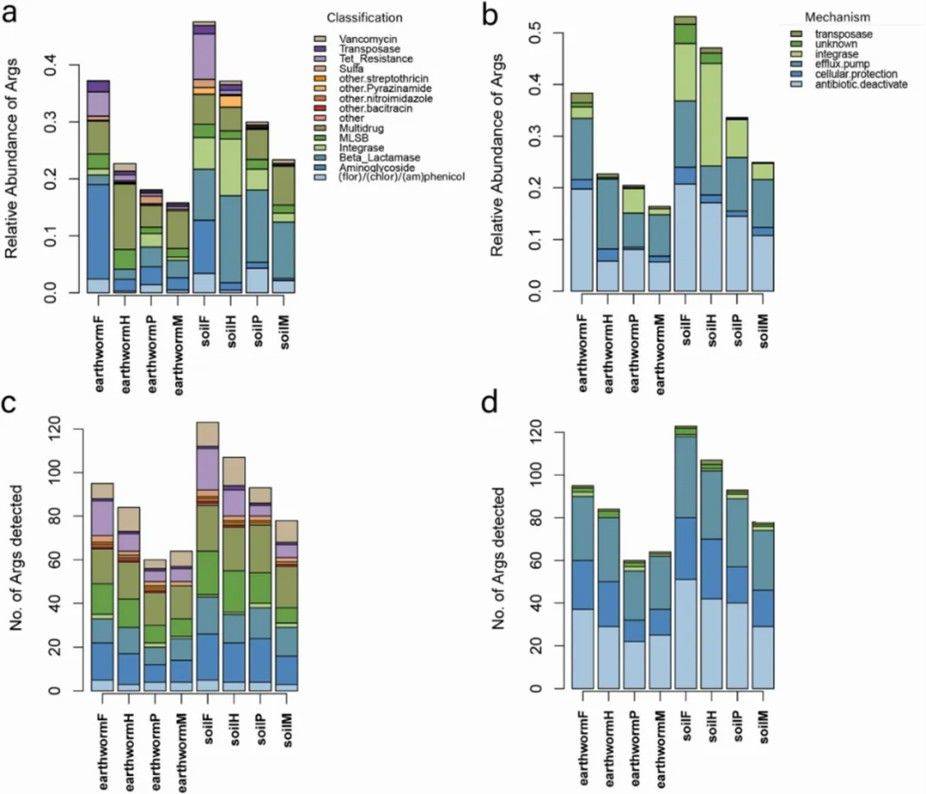 Figure 1. Relative abundance (a) and quantity (c) of ARGs in soil and earthworm microbiomes across different land use types.
Figure 1. Relative abundance (a) and quantity (c) of ARGs in soil and earthworm microbiomes across different land use types.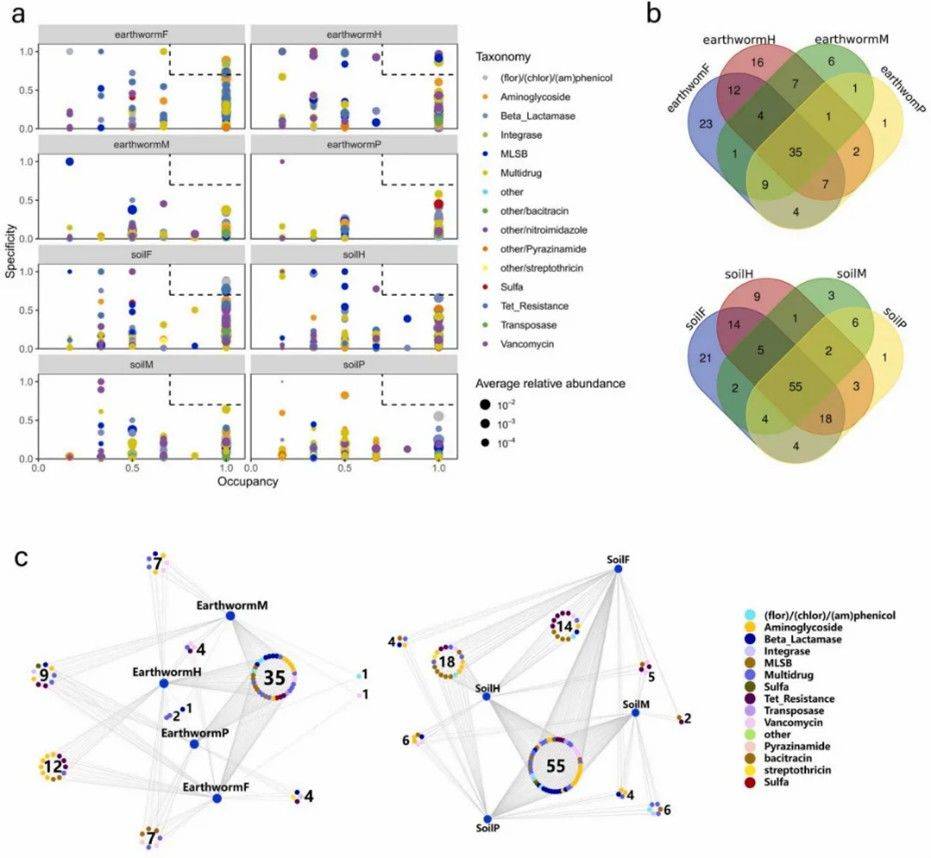 Figure 2. (a) Distribution of ARGs profiles in each habitat type; (b) Venn diagrams showing the number of shared ARGs between different earthworm and soil samples; (c) Bipartite network analysis depicting shared ARGs among different land use types in soil and earthworm microbiomes.
Figure 2. (a) Distribution of ARGs profiles in each habitat type; (b) Venn diagrams showing the number of shared ARGs between different earthworm and soil samples; (c) Bipartite network analysis depicting shared ARGs among different land use types in soil and earthworm microbiomes.