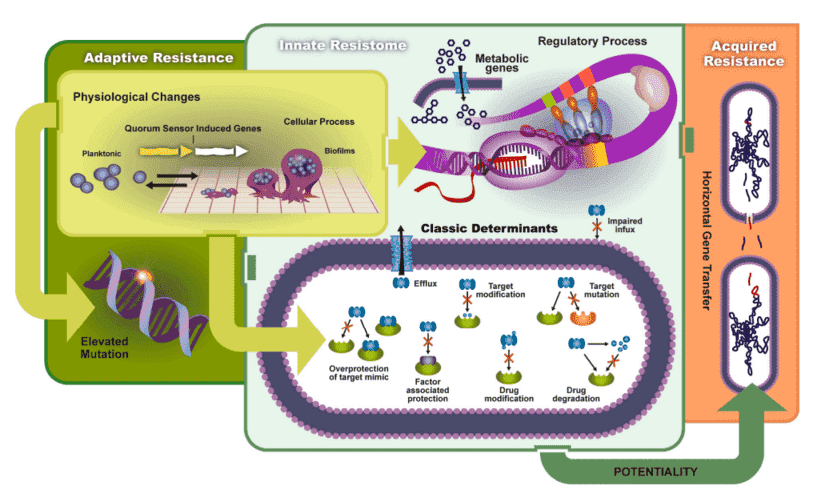
| Gene |
Classification |
Mechanism |
| catA1, catB3, cfr, etc |
(flor)/(chlor)/(am)phenicol |
deactivate |
| cmlA1, cmx(A), floR, etc |
efflux |
| qnr, etc |
- |
| aac, aacA/aphD, aacC, aacC1, aacC2, aacC4, aadA, aadD, aadE, aph, aph6ia, aphA1(akakanR), spcN-01, spcN-02, str, strA, strB, etc |
Aminoglycosides |
deactivate |
| tetA, tetB, tetC, tetD, tetE, tetG, tetH, tetJ, tetK, tetL, tetPA, tet, tetV, etc |
Tetracyclines |
efflux |
| tet(32), tet(36), tetM, tetO, tetW, tetPB, tetS, tetT, tetQ, etc |
protection |
| tet(34), tet(37), tetU-01, tetX, tet(35), etc |
- |
| GES, KPC, IMP-1, NDM-1(C), blaOXA-48, etc |
critical antimicrobial classes |
- |
| vanA, vanB, vanC, vanG, vanHB, vanHD, vanRA, vanRB, vanRC, vanRD, vanSA, vanSB, vanSC, vanTC, vanTE, vanTG, vanWB, vanWG, vanXA, vanXB, vanXD, vanYB, vanYD, etc |
enzyme targets in resistance studies |
protection |
| acrA, adeA, acrF, ceoA, cmeA, cmr, marR, mdetl1, mdtE/yhiU, mepA, mexA, mexD, mexE, mexF, mtrC, mtrD, oprD, oprJ, pmrA, qac, qacA, qacA/qacB, qacH, rarD, sdeB, tolC, ttgB, yceE/mdtG, yceL/mdtH, yidY/mdtL, ttgA, emrD, etc |
Multidrug |
efflux |
| ampC/blaDHA, ampC, bla1, blaCMY, blaCTX, blaGES, bla-L1, blaMOX/blaCMY, blaOCH, blaOKP, blaOXA1/blaOXA30, blaOXY, blaPAO, blaPER, blaPSE, blaROB, blaSFO, blaSHV-01, blaTEM, blaTLA, blaVEB, blaVIM, blaZ, cepA, cfiA, cfxA, cphA, fox5, NDM1, ampC, etc |
Beta_Lactamas |
deactivate |
| mecA, pbp, pbp2x, Pbp5, penA, etc |
protection |
| ereB, lnuA, lnuB, lnuC, mphA, mphB, mphC, vatB, vatC, vatE, vgb, vgbB, etc |
MLSB |
deactivate |
| carB, ImrA, matA/mel, mdtA, mefA, msrC, oleC, vgaA, vgbB, msrA, etc |
efflux |
| erm, ermA, ermA/ermTR, ermB, ermC, ermF, ermJ/ermD, ermK, ermT, ermX, ermY, etc |
protection |
| dfrA, folA, etc |
Sulfonamides |
deactivate |
| sul, etc |
protection |
| IS613, tnpA, Tp614, etc |
MGEs |
transposase |
| int, etc |
integrase |



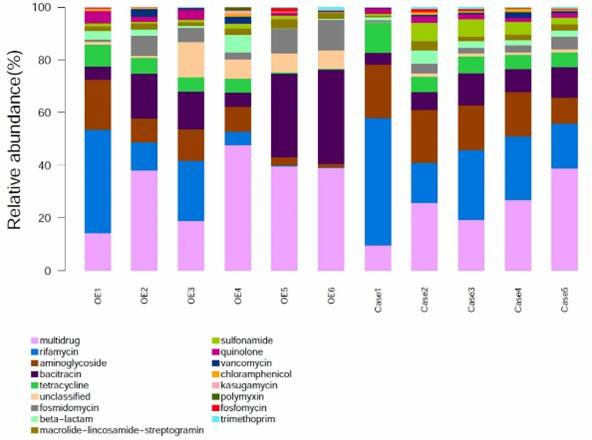
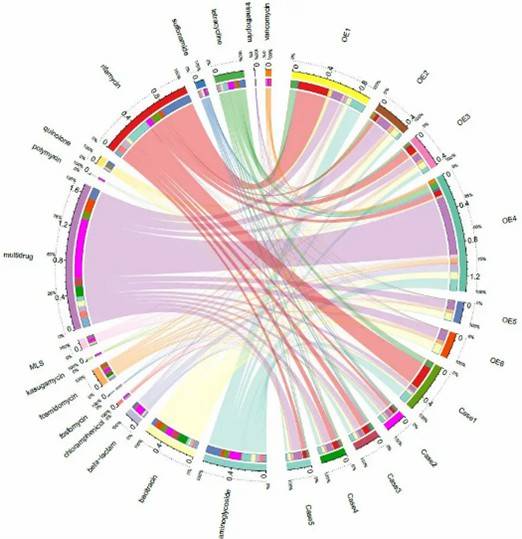
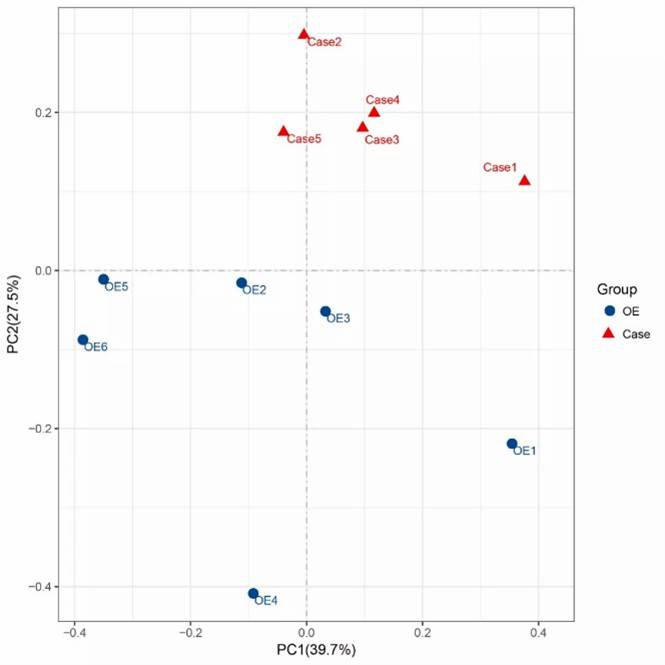
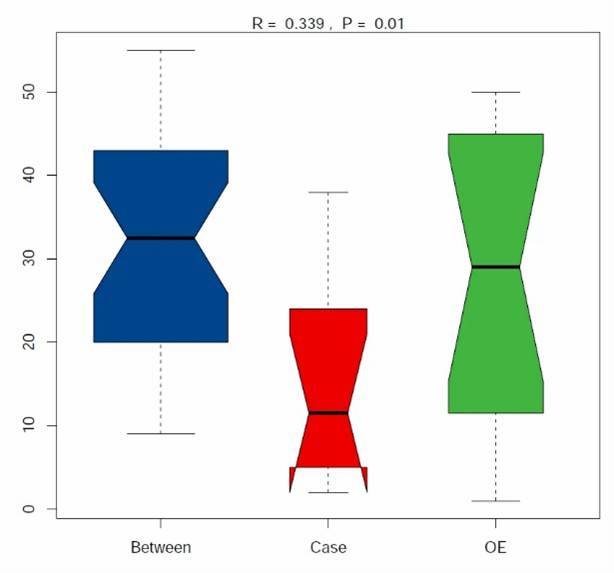
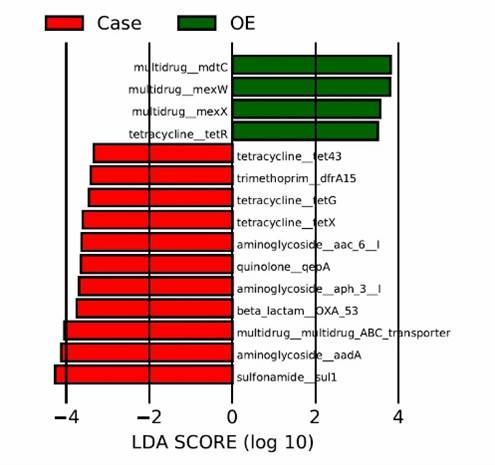
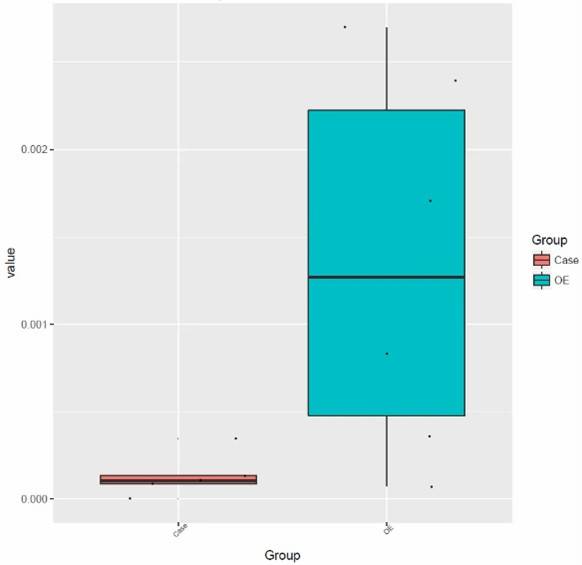

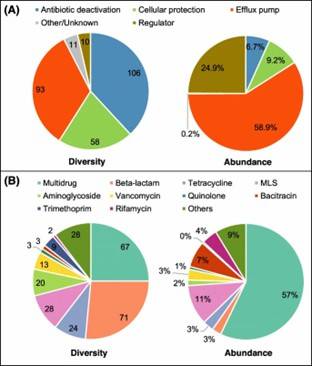 Fig 1. Composition of ARGs and regulator genes in 26 soil metagenomes.
Fig 1. Composition of ARGs and regulator genes in 26 soil metagenomes.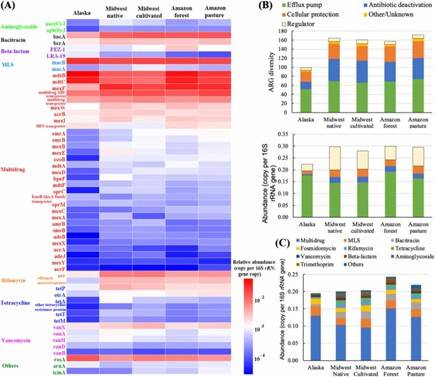 Fig 2. Diversity and abundance and of ARGs among soils of three ecosystems.
Fig 2. Diversity and abundance and of ARGs among soils of three ecosystems.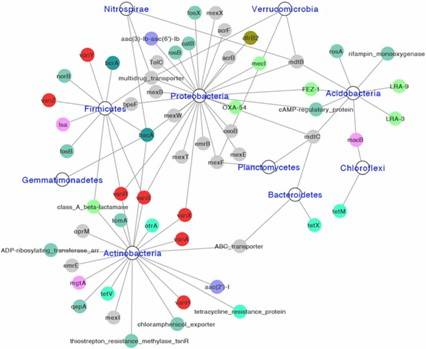 Fig 3. Network showing identified hosts of ARGs at phylum level. Different colors represent different classes of ARGs
Fig 3. Network showing identified hosts of ARGs at phylum level. Different colors represent different classes of ARGs