The Importance of Oral Microbiome
The microbiome found in the mouth is highly complex with more than 700 species, including viruses, fungi, bacteria, archaea, and protozoa, some of which are primarily disease-associated. Under normal conditions, the interaction between oral microbiome, and between microbiome and host maintains a dynamic balance, but oral homeostasis is easily disturbed by external factors. The ecological imbalance of oral microorganisms can not only induce a variety of oral diseases (such as caries, periapical disease of dental pulp, periodontal disease, etc.), but are also closely related to tumor, diabetes, rheumatoid arthritis, cardiovascular disease, premature delivery and other systemic diseases, which have a great impact on human health.
Accelerate Research and Practice in Oral Microbiome
We are equipped with Illumina Hiseq/Miseq, Ion PGM, PacBio RSII, and Nanopore MinION platforms to help you identify the microorganisms presented in the oral cavity, and analyze the diversity, abundance, evolutionary relationship, and disease implications of the oral microbiome. Besides, we also provide qualitative and quantitative studies of the oral microbiome using real-time qPCR.
The Main Directions
- Study the differences of oral microbial in the composition and abundance between the diseased group and healthy group
- Explore and compare the differences of oral microbial community structures among individuals at different intervention stages
- Study changes in the oral microbiome at different developmental stages
- Search for biomarkers or drug targets of certain diseases
What Can We Do?
- 1. Identification of oral microorganisms
- 2. Quantitation of oral microorganisms
- 3. Study the evolutionary relationship of oral microorganisms
- 4. Study the relationship between oral microorganisms and diseases
Note: Our service is for research use only, and not for therapeutic or diagnostic use.
Detectable Objects
DNA samples of oral microbiome from humans and animals (such as mouse rat, sheep, monkey, horse, fish, etc.) can be tested.
Detectable Microorganisms
Viruses, fungi, bacteria, archaea, etc.
Technical Platforms
We are equipped with Illumina HiSeq/MiSeq, Ion PGM, PacBio SMRT systems, Nanopore systems, PCR-DGGE (PCR-denaturing gradient gel electrophoresis), real-time qPCR, clone library, and other detection platforms.
Sample Requirement
-

- DNA sample: DNA ≥ 500ng, concentration ≥ 10ng/ul, and OD260/280 = 1.8-2.0.
- Ensure that the DNA is intact and non-degraded, avoiding repeated freezing and thawing cycles.
- Dry ice or ice packs shall be used for sample delivery.
- We provide superior MicroCollect™ oral sample collection products for the collection of oral microbiome from saliva or oral cells.
Workflow
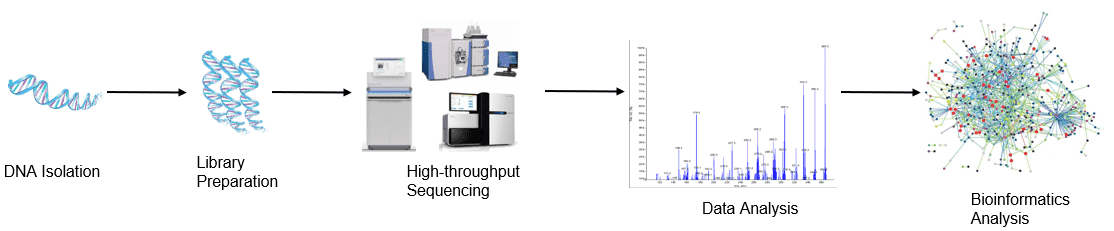 Figure 1. High throughput sequencing analysis process
Figure 1. High throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process
Bioinformatics Analysis
| OTU Clustering | Distribution-Based OTU-Calling |
| Rarefaction Curve | |
| Shannon-Wiener Curve | |
| Rank Abundance Curve | |
| Diversity Index | |
| OTU-Based Analysis | Heatmap |
| VENN | |
| Principal Components Analysis (PCA) | |
| Hierarchical Clustering | |
| Comparative Analysis | RDA/CCA |
| PCA/PCoA | |
| Network Analysis | |
| Functional analysis | BLASTX, KEGG, eggNOG, CAZy, CARD, ARDB etc. |
| PICRUSt | |
| TAX4Fun | |
| Phylogenetic Analysis | (Un)Weighted Unifrac |
| Phylogenetic Trees |