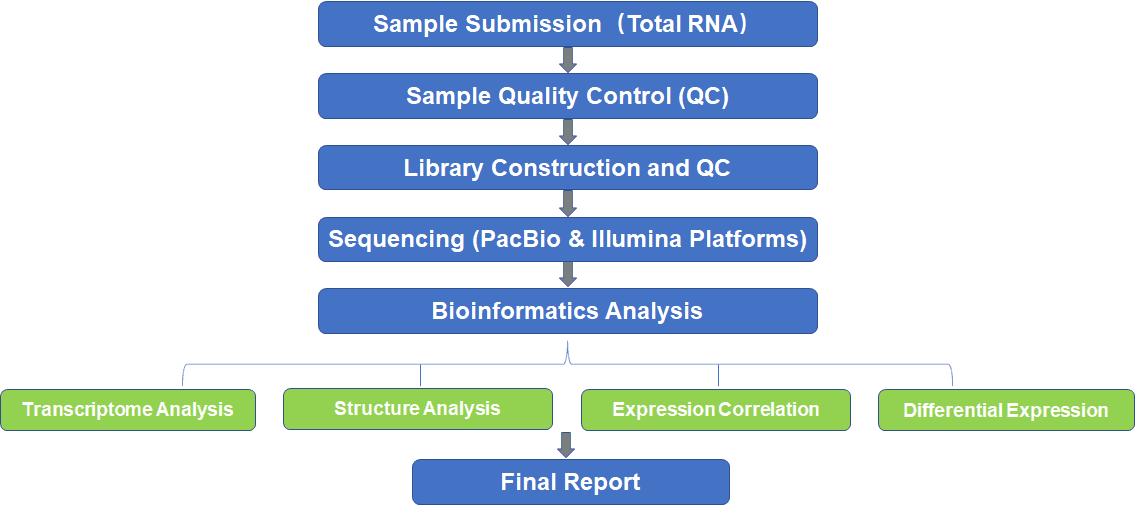
Our fungal iso-seq or bacterial iso-seq can offer complete solutions for full-length transcript sequencing. It is a powerful technique using combined Illumina and PacBio SMRT sequencing platforms to identify both high and low expressors in a single bacterial or microbial population. PacBio SMRT sequencing enables the sequencing of contiguous full-length transcripts and captures each transcript's start, splice sites, and polyadenylation from a single read. Illumina short reads are further mapped to the PacBio SMRT full-length transcripts for more accurate quantification analysis and error correction.
Microbial iso-seq is becoming increasingly important for basic research. It accurately characterizes the diverse landscape of transcript isoforms, further providing key information on the functional biology of genomes for developing potential applications of these abundant microbes in disease, medicine, agriculture, and environment, for example, predicting resistance to specific antibiotics, quantifying gene expression changes, understanding host-pathogen immune interactions, and tracking disease progression.