Next-Generation Sequencing of Bacterial Genomes: Introductions, Technologies, and Applications
Inquiry >Bacteriology studies multiple aspects of bacteria including morphology, metabolism, function, and ecology. The approaches generally involve identification, classification, and characterization. The development of next-generation sequencing (NGS) technologies allows researchers to investigate microorganisms from a broader and deeper perspective, enabling better characterization of the bacterial genome as well as deeper taxonomic identification of complex microbiomes in specific environments such as soil, ocean, and even human body. NGS is capable of processing multiple DNA sequences in parallel and dramatically reduces sequencing cost and time. The technical developments, applications, and research of NGS are moving forward rapidly over the years. Many studies have highlighted the use of NGS strategies in bacterial classification, microbiome, comparative genomics, and evolutionary genomics, etc.
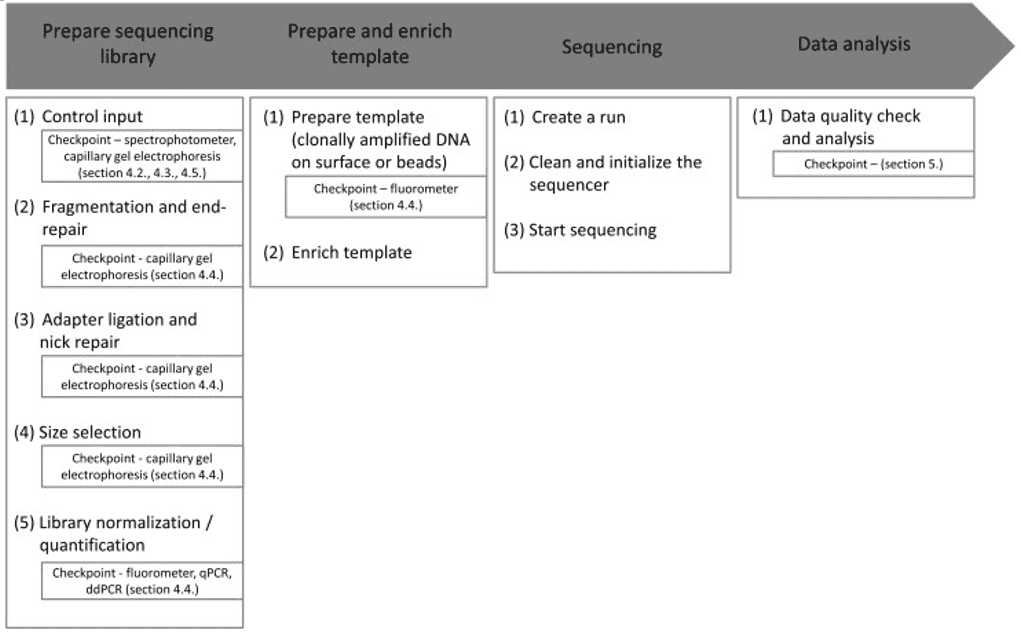
16S rRNA amplicon sequencing, metagenomics, and metatranscriptomics are widely used NGS strategies in bacterial identification and characterization. The key features of NGS include in vitro library construction and clonal amplification, array-based sequencing enables DNA fragments to be multiplexed, and DNA solid-phase fixation. Compared with Sanger sequencing based on selective binding chain-terminating dideoxynucleotides during in vitro replication by DNA polymerase, NGS has high sequencing depth, which allows for high sensitivity and comprehensive genomic coverage. More significantly, NGS allows for higher sample throughput and can produce massive sequencing data in a single run.
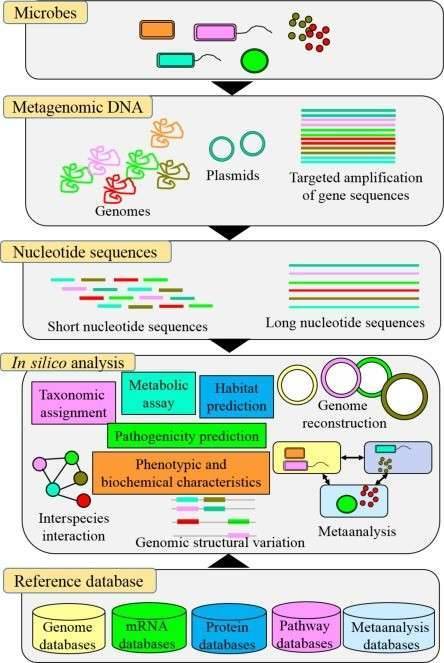
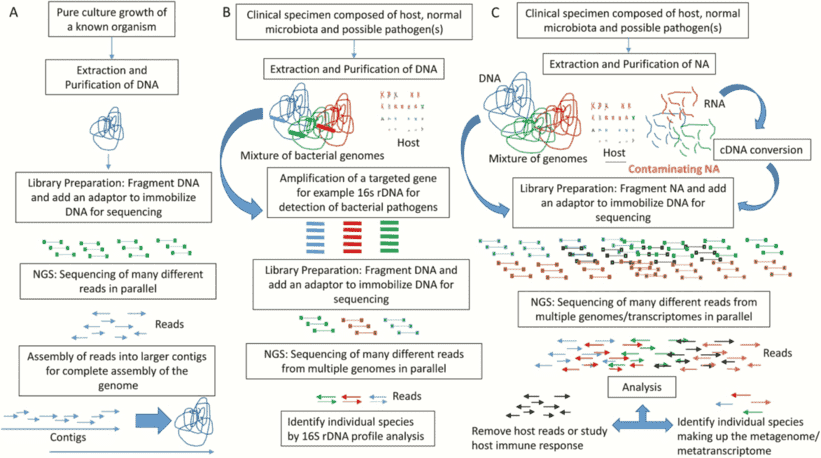 Figure 1. An overview of the different applications of next-generation sequencing analysis. (Simner 2017)
Figure 1. An overview of the different applications of next-generation sequencing analysis. (Simner 2017)
Amplicon Sequencing
16S ribosomal RNA sequencing has been used extensively in bacterial phylogeny and the taxonomic composition of a microbiome. With the advent of NGS technology, it has become easier and more economical to produce 16S rRNA gene sequences in mixed complex samples without culturing steps, allowing rapid identification of bacteria of interest. There are several high-throughput sequencing technologies that fulfill the versatile needs of bacteriology projects, such as PacBio SMRT Sequencing and Nanopore Sequencing, which both provide full-length 16S sequencing and guarantee accurate outcomes.
Whole Genome Sequencing
Whole genome sequencing is an unbiased approach for bacterial profiling. Sequencing the entire bacterial genome is important for generating accurate reference genomes, microbial identification, and other comparative studies. Microbial whole-genome de novo sequencing is an important tool for mapping genomes of novel organisms, whereas microbial whole-genome resequencing investigates genomes of known organisms, or comparing genomes across multiple samples. With various NGS platforms (such as PacBio SMRT and Illumina HiSeq PE150) of different features and merits, WGS can be performed to satisfy specific needs.
Shotgun Metagenomic Sequencing
Shotgun metagenomic sequencing overcomes many limitations of the amplicon sequencing. It generates billions of DNA base pairs of all organisms present in a given complex sample without bias. This method allows researchers to detect the abundance of microbes and investigate bacterial diversity in a variety of environments. With the ability to capture the whole picture of microbial genetic information, metagenomic sequencing also provides the approach to understand the functions encoded by the genomes of the microbiome, which contains bacteria, archaea, and fungi. Shotgun metagenomic sequencing produces vast amounts of data that can be analyzed by powerful computing technologies.
Bacterial Metatranscriptome Sequencing
Metatranscriptomics offers a more specific perspective than metagenomics because it can reveal details about bacterial populations with transcriptional activity, rather than just identifying the genetic content as in the metagenomic analysis. Metatranscriptomics studies the functions and activities of the complete set of transcripts (RNA-seq) from environmental samples. Metatranscriptome sequencing has provided significant insights about the microbial community structure and functional characterization of the complex metabolic interactions within a single microbiome, which help to elucidate key genes and pathways, discover gene activity diversity, and conduct gene expression comparison.
References
- Simner PJ; et al. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clinical Infectious Diseases. 2017, 66(5):778-88.
- Cao Y; et al. A Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Frontiers in Microbiology. 2017, 8(1829).