Introduction to Metagenomics
Lately, metagenomics or environmental genomics has become a strong method for gathering microbial community data, bypassing individual species cultivation. Conventional metagenomic sequencing, nevertheless, typically involves high costs and time. Therefore, only scant info about highly complicated natural populations, such as those that populate soils and lake sediments, can be collected.
Metagenomics has reshaped the research of natural bacterial diversity and supplied a wealth of biological insight by sequencing total microbial DNA collected from an environmental area. The complexity of reconfiguring short sequence fragments inferred from myriad organisms in society into well-resolved biological entities, like bacterial communities, viruses, and plasmids, however, stays a key obstacle that limits the usefulness of this strategy. In this issue, for the interpretation of metagenomic data at the stage of individual genomes, an effective model is employed without depending on reference sequences of cultured isolates that can be produced for only a mere portion of microbial life. They present a new process for leveraging the fact that genes co-vary in abundance in the metagenome in physical bonding, such as on the same chromosome. Even in the complicated context of the human gut, their process helps identify the purpose of bacterial diversity and brings people to the true objective of a full, high-resolution description of the bacterial population in environmental samples.
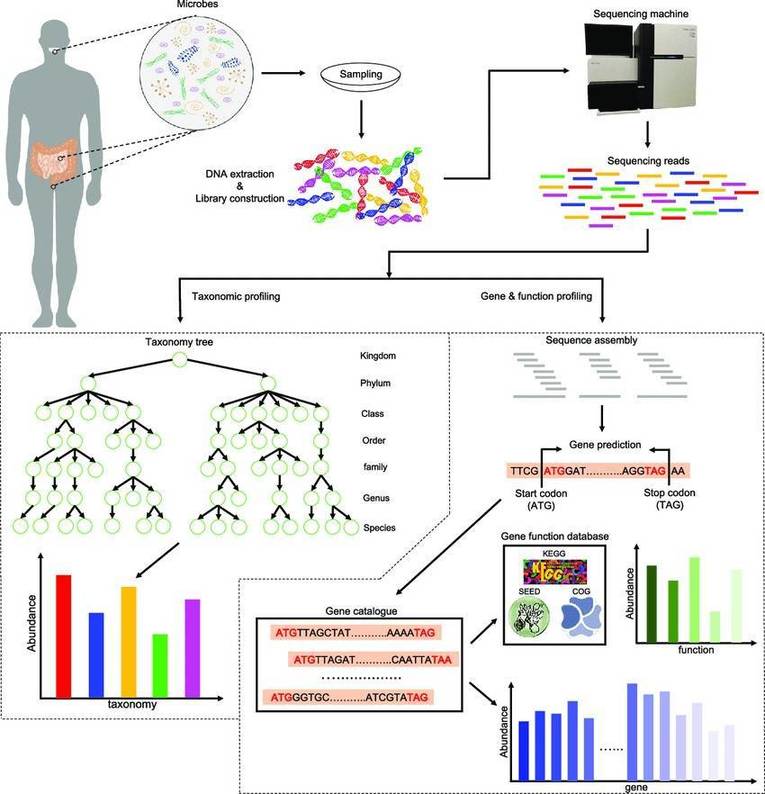
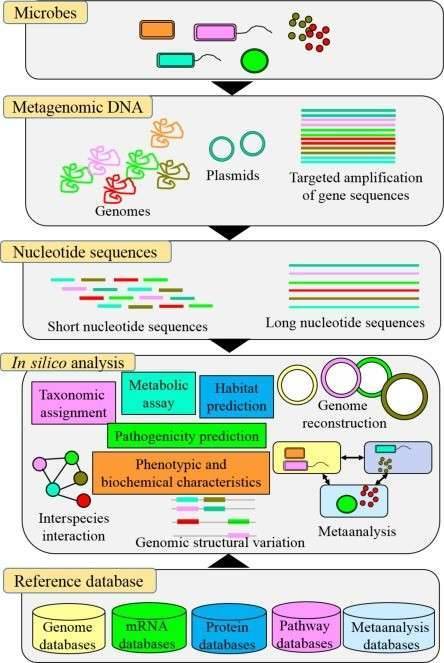 Figure 1. Schematic presentation of metagenomic and bioinformatic analyses of the microbial community in atypical environment. (Dash, 2018)
Figure 1. Schematic presentation of metagenomic and bioinformatic analyses of the microbial community in atypical environment. (Dash, 2018)
Metagenomics Technology
The accruing amount of DNA data sets has resulted in metagenomics innovation, which enables us to acquire diverse gene or pathway elements from the group. To increase our understanding of microbial ecology, these extended genome sequences are currently being manipulated for novel biotechnological applications. To know the framework (gene/species richness and distribution) and the functional (metabolic) possibilities of environmental bacterial diversity, metagenomics methods have been taken.
Metagenomic research is grouped into three classes depending on various screening methods: (i) shotgun analysis using mass genome sequencing, (ii) activity-driven findings intended to look for microbial features, and (iii) sequence-driven experiments that connect genome data with genes of concern in phylogenetic or functional markers. More lately, next-generation sequencing techniques have been established to straightforwardly evaluate the entire compilation of genes without creating a metagenomic library within an environmental data set.
According to their random and directed sequencing strategies, the metagenomics methods can be defined as unselective (shotgun analysis and next-generation sequencing) and targeted (activity-driven and sequence-driven studies) metagenomics. Due to its simplification and price in DNA sequencing, unselective metagenomics is becoming an incredibly popular technique. Many microbial community random sequencing ventures, like bacteria, archaea, and viruses, have been confirmed.
Targeted Metagenomics Strategies
It is feasible to use two distinct metagenomics procedures: unselective and targeted metagenomics sequencing of an environmental DNA pool is becoming incredibly popular using high-throughput sequencing innovation. Even though in the initial phases of development, techniques for creating and analyzing metagenomics data persist. An intentionally selected DNA pool is confined to sequencing in a targeted metagenomics method to decrease genetic complexity. Typically, the selection process is centered on (i) sequence-driven or (ii) function-driven screening. Targeted metagenomics can offer wide coverage and substantial duplication of sequences for aimed genes by concentrating attempts on sequence evaluation and unveiling genome regions directly connected to an ecological process, even at low abundances within a metagenome. For genome arrangement and successive data assessment, a greater sequence range of the acquired target metagenomics can be advantageous.
Targeted metagenomics based on sequence-driven screening
One of the most popular methods of identifying genome fragments inferred from factions of microorganisms that have not yet been cultivated or that play an essential part in the ecosystem is the use of the 16S ribosomal RNA gene (16S rDNA) as a phylogenetic marker gene.
Targeted metagenomics based on functional-driven screening
As a substitute to phylogenetic markers, a more immediate path to the finding of gene clusters with associated metabolic functions in bacterial diversity is the application of known gene functions of concern. In addition, function-driven testing methods can possibly offer a way of revealing unexplored genes or gene families that cannot be discovered by sequence-driven methods, although screening processes are often needed.
References
- Nath H, Mallick A, Roy S, et al. Computational modelling supports that dengue virus envelope antibodies can bind to SARS-CoV-2 receptor binding sites: Is pre-exposure to dengue virus protective against COVID-19 severity?. Computational and Structural Biotechnology Journal. 2021 Jan 1;19.
- Dash HR, Das S. Molecular methods for studying microorganisms from atypical environments. Methods in Microbiology. 2018 Jan 1;45.
- Mick E, Sorek R. High-resolution metagenomics. Nature biotechnology. 2014 Aug;32(8).