Overview of Microbial Detection and Identification
The detection and identification of microbes constitute a salient dimension in microbiological studies, with applications spanning diverse sectors, namely environmental science, industry, and medicine. The pivotal role and expansive application of microbial detection and identification rest in safeguarding public health, securing food safety, promoting environmental protection, and enhancing medical diagnostics. Through accurate and expedient microbial detection and identification, it is feasible to effectively inhibit disease transmission, maintain environmental health, improve food quality and production efficiency, and foster human health and societal advancement.
The methodologies for microbial detection and identification have continually evolved, progressing from traditional, time and labor-intensive techniques, such as cultivation, morphology, physiology, pathology, and biochemical tests, to contemporary spectrometry and genetic technologies. In recent years, the speed of advancement in microbiology research has been remarkable. Microbial identification methods can be categorized into two types: phenotypic and genotypic. Phenotypic methods elucidate microbes based on their phenotype attributes, while genotypic methods depend on the interpretation of genetic information.
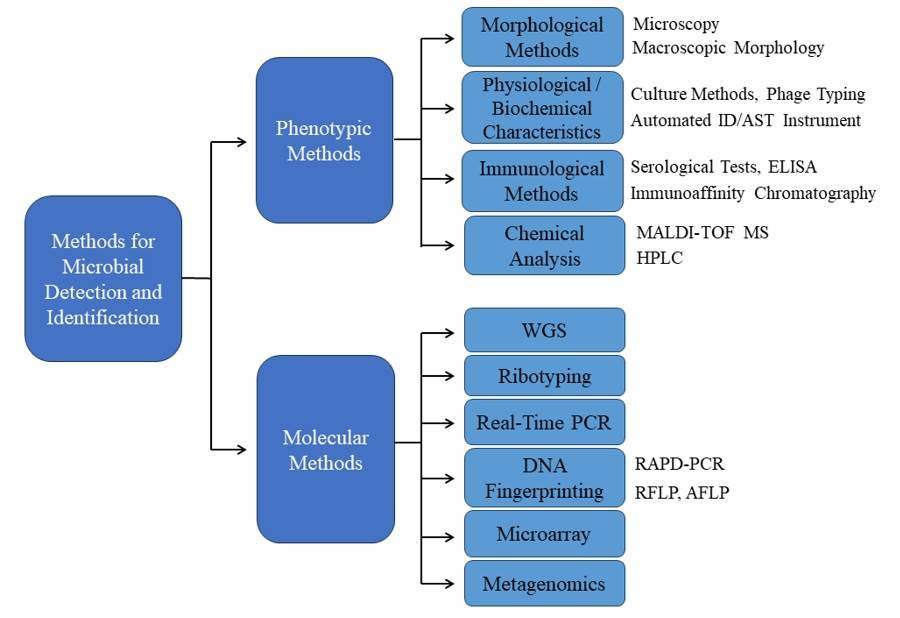 Figure1. Methods for Microbial Detection and Identification.
Figure1. Methods for Microbial Detection and Identification.
Phenotypic Methods
Morphological Methods
- Microscopy
Microscopic microbial identification hinges upon a repertoire of microscopy techniques, encompassing optical, electron, and scanning probe microscopy, which serve to unveil microorganisms imperceptible to the unaided eye. The synergy between these methodologies and a profound understanding of microbiology expedites the retrieval of pivotal morphological data essential for microbial identification. For instance, Nemati et al. (2016) employed optical microscopy to meticulously scrutinize the morphological traits of bacteria and fungi present in pharmaceutical and food samples, thereby facilitating their accurate identification and classification.
- Macroscopic Morphology
The cultivation of microorganisms continues to be an essential methodology within the field of microbial research. This is predominantly due to the fact that divergent microbial species present unique colony morphologies. The meticulous inspection of such colony morphologies supports a preliminary identification of the microorganisms, founded on their macroscopic characteristics. Diba et al. conducted a study to assess the efficacy of morphological methods, encompassing macroscopic and microscopic traits, in discerning Aspergillus species from both environmental and clinical origins. Employing four distinct differential media, they scrutinized colony characteristics vis-a-vis standard Aspergillus strains. Parameters examined included colony diameter, color, exudates, and texture, alongside microscopic traits like conidial heads, stipes, vesicle morphology, and conidia dimensions. Findings revealed that morphological techniques employing the specified media successfully discriminated 11 Aspergillus species, with A. flavus, A. niger, and A. fumigatus predominating among isolates. The investigation concluded that morphological methodologies, when coupled with differential media, offer reliability and sensitivity in identifying clinically relevant Aspergillus species.
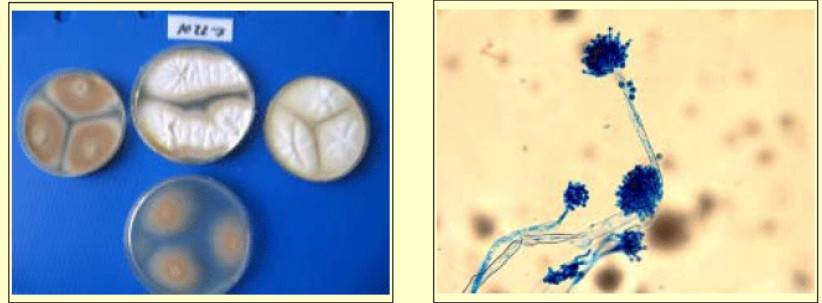 Figure 2. Colony feature of A. tereus in four differential media. (Diba et al., 2007)
Figure 2. Colony feature of A. tereus in four differential media. (Diba et al., 2007)
Physiological / Biochemical Characteristics
- Culture Methods
Culture methods involve assessing the growth characteristics and morphology of microorganisms in different culture media. Pure cultures obtained under various conditions provide valuable biochemical clues for microbial detection and identification. In the study by Nemati et al. (2016), cultural characteristics, such as growth patterns and biochemical reactions, were utilized for the identification of microbial contaminants in pharmaceutical and food samples.
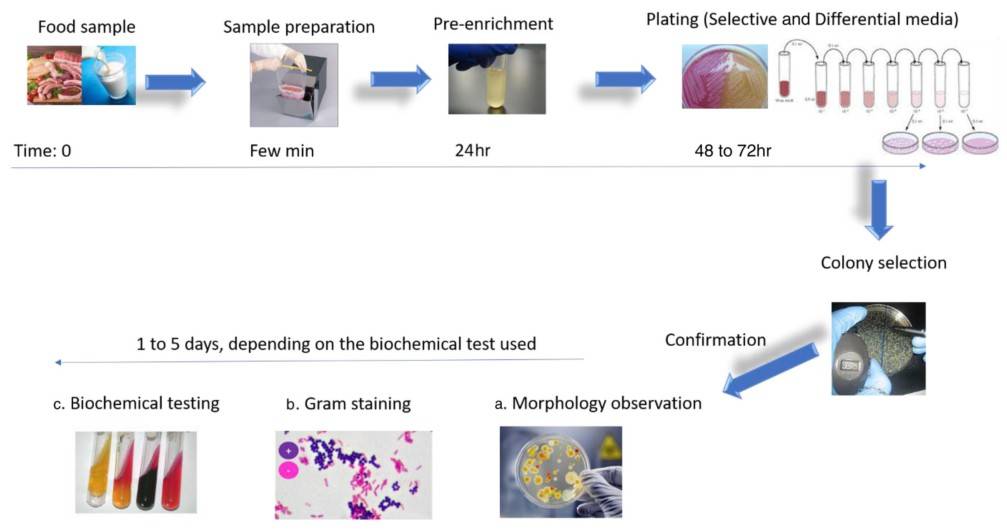 Figure 3. Typical workflow for microbial identification according to the conventional culturing and plating methodologies. (Ferone et al., 2020)
Figure 3. Typical workflow for microbial identification according to the conventional culturing and plating methodologies. (Ferone et al., 2020)
- Phage Typing
The fundamental and indispensable technique of utilizing phage typing to recognize individual bacterial strains and trace the origin of infectious outbreaks sheds light on the intricacies of bacterial genetics and disease transmission. Phage typing harnesses bacteriophages – viruses that infect bacteria – to specify unique strains within a bacterial species, basing this differentiation on their respective susceptibility patterns. Lee et al. conducted a study aimed at characterizing 90 Methicillin-resistant Staphylococcus aureus (MRSA) isolates obtained from Kyung Hee University Hospital in Korea. Through the use of bacteriophage typing, pulsed-field gel electrophoresis (PFGE), and antimicrobial susceptibility testing, the research revealed that 80% of the strains could be phage-typed. Phage type 83A emerged as the most prevalent, followed by phage type D11. PFGE analysis delineated two major clusters, representing lyso group III strains and those typable with regional additional phages. The study emphasized the notable resistance rates observed against ciprofloxacin, erythromycin, tetracycline, gentamicin, and clindamycin, while no strains displayed intermediate resistance to vancomycin or resistance to mupirocin. These findings underscore the complementary nature of phage typing and PFGE in the comprehensive characterization of MRSA strains.
Automated ID/AST Instrument
Modern microbiology instruments offer rapid and accurate microbial identification and susceptibility testing. These instruments interpret biochemical profiles and match them against databases, significantly reducing the time required for testing. Cherkaoui et al. explore the transformative potential of Total Laboratory Automation (TLA) amidst the increasing complexity and costs of diagnostic bacteriological testing. Their study underscores that TLA goes beyond mere replication of manual procedures, requiring a fundamental shift in laboratory workflows, particularly impacting pathogen identification and antimicrobial susceptibility testing. Highlighting efficiency improvements, the research introduces a fully automated solution for Antimicrobial Susceptibility Testing (AST) via disk diffusion testing, with evidence suggesting positive clinical outcomes. TLA's capability to manage diverse culture-based testing procedures is demonstrated, offering benefits such as enhanced productivity, traceability, and reduced Turnaround Time (TAT). While the integration of artificial intelligence (AI) holds promise for further workflow enhancements, validation studies are imperative for the automatic processing of negative cultures. TLA systems, exemplified by the Colibri coupled to the Radian, facilitate the implementation of EUCAST rapid AST, potentially enhancing patient outcomes.
 Figure 4. Total Lab automation workflows. (Cherkaoui et al., 2022)
Figure 4. Total Lab automation workflows. (Cherkaoui et al., 2022)
Immunological Methods
- Serological Tests
Serological testing is a valuable approach utilized in microbial detection and identification, facilitating the categorization of similar species or strains within the same species by detecting microbial antigens and antibodies. This method involves performing serological assays specific to the targeted microorganism, utilizing antisera derived from known bacterial strains or types.
- ELISA
Enzyme-linked immunosorbent assay (ELISA) is a versatile analytical technique used to detect the presence of microbial antigens or antibodies in a given sample. ELISA has extensive applications in clinical diagnosis, pathological studies, and quality control assessments. This method allows for the detection of either antigens or antibodies, enabling the identification of different microbial strains simultaneously and characterizing the epitope distribution on microbial surfaces. In their academic article, Zhao and colleagues delve into the historical inception of Enzyme-Linked Immunosorbent Assay (ELISA), as pioneered by Engvall and Perlmann in 1971, granting particular emphasis to its inherent limitations. Specifically, the researchers underscore ELISA's characteristic moderate sensitivity and dependence on enzymatic activity, hinting at the potential drawbacks of these features. The paper then segues into the auspicious possibilities presented by the advent of nanotechnology. The authors introduce the concept of synthesizing innovative nanomaterials loaded with unique properties, advocating this as a promising solution to the aforementioned limitations of traditional ELISA. They subsequently underscore the significant enhancement of ELISA's sensitivity and the simplicity of its operations attributable to the incorporation of nanomaterials. Focusing on the compelling diversity in signal detection modes, Zhao et al. present a comprehensive exploration of an array of methods ranging from colorimetric to fluorescent, electrochemical, photothermal, and Raman scattering sensing modalities. They articulate the necessity of conducting structured and systematic reviews within this academic sphere, as a preventive measure against duplicative research efforts, ultimately fueling the progressive development of effective, pragmatic ELISA techniques. In conclusion, the article delivers an overview of the impressive advancements being made in the realm of nanomaterial-based ELISAs, while additionally outlining the potential repercussions these developments stand to impart on clinical diagnostics, environmental monitoring, and food safety measures.
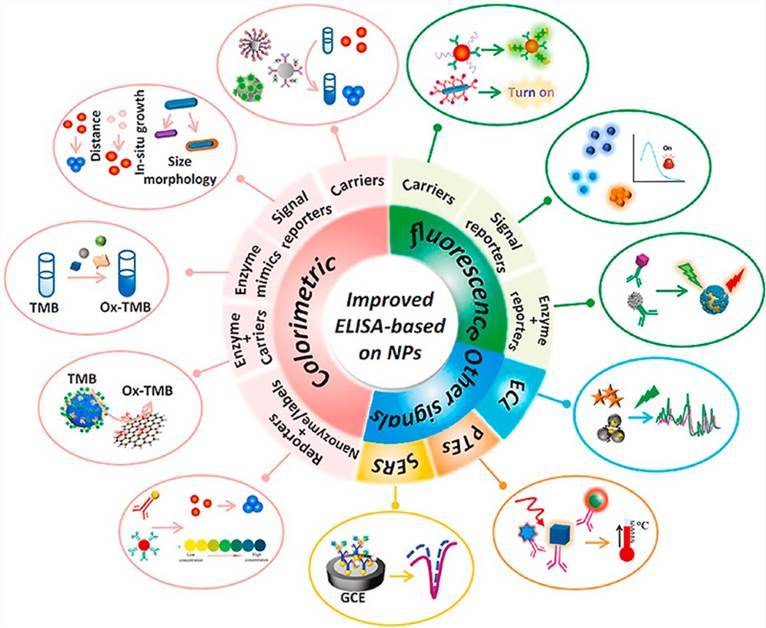 Figure 5. Recent improvements in enzyme-linked immunosorbent assays based on nanomaterials. (Zhao et al., 2021)
Figure 5. Recent improvements in enzyme-linked immunosorbent assays based on nanomaterials. (Zhao et al., 2021)
Immunoaffinity Chromatography
Immunoaffinity chromatography (IAC) integrates liquid chromatography (LC) with the specificity of antibodies or related reagents. This innovative methodology involves coupling antibodies with a solid inert matrix, typically within a column structure, to selectively capture microbial antigens. The captured antigens are then separated from other materials using a series of buffer solutions, ensuring purification and isolation.
Chemical Analysis
- MALDI-TOF MS
Matrix-Assisted Laser Desorption / Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) is a proteomics-based approach for microbial identification and characterization, which has been widely used as an identification technique in microbiology laboratories. With significant advantages compared with other tools, such as biochemical identification and phenotypic tests, MALDI-TOF MS revolutionized the approach to the rapid and accurate identification of bacteria, shortens the period from days to one hour within.
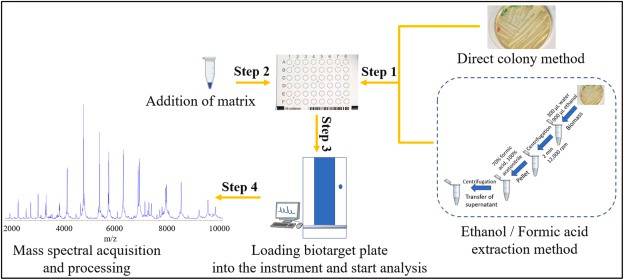 Figure 6. Workflow for bacterial identification using MALDI-TOF MS. (Ashfaq et al., 2022)
Figure 6. Workflow for bacterial identification using MALDI-TOF MS. (Ashfaq et al., 2022)
- HPLC
Identification of microorganisms by means of high-performance liquid chromatography (HPLC) has provided an alternative to diversified approaches, which can target multiple microbial components such as cell wall mycotic acid, nucleotides, and so on.
Molecular Methods
Whole-Genome Sequencing
Whole-genome sequencing provides a comprehensive understanding of the entire genome, which not only determines the likelihood that genetic material comes from an individual or group but also suggests additional information on genetic relationships, origin or susceptibility to specific diseases. The major processes of whole-genome sequencing workflow are alignment, variant calling, annotation and calculating related metrics. Hasman et al.'s study outlines the practicality of utilizing WGS directly on clinical specimens as an innovative tool in clinical microbiology, aiming to lessen diagnostic delays thereby improving treatment and control measures. The study was carried out using thirty-five arbitrary urine samples collected from patients suspected of urinary tract infections. Bacterial cultures were procured from nineteen samples, with only seventeen providing pure cultures. The investigation comprised conventional microbiology, WGS of isolated bacteria, and direct sequencing on urine sample pellets. A rapid mode of analyzing the acquired sequencing data was also developed. WGS enriched the identification of cultivated bacteria, and substantial concurrence was observed between phenotypic and forecasted antimicrobial susceptibilities. Upon contrasting the results of WGS of cultured isolates and urine sample sequencing directly, there was total agreement in species identification, multilocus sequence typing, and phylogenetic relationships for Escherichia coli and Enterococcus faecalis isolates. Moreover, direct sequencing from the urine facilitated bacterial identification in polymicrobial samples and identified potential pathogenic strains in some culture-negative samples. Findings suggest that applying WGS directly on clinical samples can provide clinically relevant information while significantly reducing diagnostic times. However, the need for data interpretation remains a hindrance to its clinical implementation. To manage this issue, a publicly accessible bioinformatic tool was developed and introduced as part of this investigation.
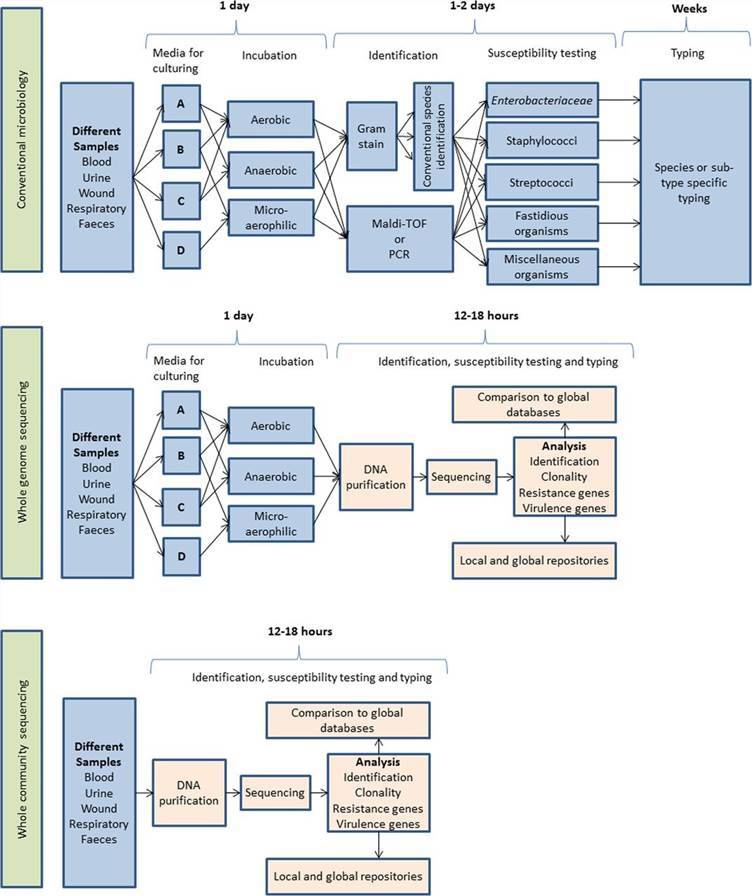 Figure 7. Schematic representation of the workflow anticipated after adoption of whole-genome sequencing used either on cultured isolates or directly on the clinical samples, with an expected time scale. (Hasman et al., 2014)
Figure 7. Schematic representation of the workflow anticipated after adoption of whole-genome sequencing used either on cultured isolates or directly on the clinical samples, with an expected time scale. (Hasman et al., 2014)
Ribotyping
Ribotyping is a methodology that uses systematics-based analysis of rRNA for bacterial identification and characterization. Given the high conservation within bacterial species of rRNA genes (such as 16S rRNA), discerning the polymorphisms of 16S rRNA gene elucidates the evolutionary spectrum of bacterial species. This, in turn, provides clues for bacterial taxonomy, classification, epidemiological investigations, and population biology. Salloum et al. present an investigation delving into the utilization of ribotyping methodology for the identification of Legionella species, a task known to pose complexities through conventional means. The ribotyping approach integrated the application of the enzyme HindIII alongside an innovative probe designated as OligoMix5. Within this study, the methodology was deployed across a cohort of 123 Legionella strains, encompassing 78 type or reference strains spanning 44 distinct species, alongside eight clinical and 37 environmental isolates. The versatility and efficacy of the ribotyping technique were convincingly demonstrated through the successful species-level identification of all 123 Legionella isolates. Specifically, concerning Legionella pneumophila, comprising 15 known serogroups, the method adeptly delineated eight discernible patterns. To ascertain the resilience of their findings, researchers additionally subjected 45 field strains to two alternative analyses – the randomly amplified polymorphic DNA (RAPD) technique and the intergenic 16S-23S ribosomal spacer PCR analysis (ITS 16-23S). Subsequently, every strain proved identifiable through the amalgamated methodologies. However, the ribotyping approach exhibited superior efficiency and effectiveness compared to its counterparts. These findings underscore the significant potential of ribotyping methodology for routine identification and diagnosis of Legionella species.
Real-Time PCR
The real-time Polymerase Chain Reaction test, also known as quantitative PCR (qPCR), is a procedure employed for the amplification, simultaneous monitoring, quantification or semi-quantification of target DNA molecules. It serves as a conventional technique in biological laboratories that provides insights into microbial gene expression and phenotypes at the genetic level. Haakensen et al. detail the development of a real-time multiplex PCR to differentiate between beer-spoilage Firmicutes and non-Firmicutes bacteria—typically environmental contaminants of innocuous nature. They honed in on a part of the 16S rRNA gene that is strongly conserved within, and nearly exclusive to, the Firmicutes phylum. They proved that the real-time PCR assay, employing a hydrolysis probe targeting this gene region, effectively detects ten Firmicutes genera, known to compromise beer, while avoiding interference from twelve out of thirteen non-Firmicutes genera occasionally found in beer. The only non-Firmicutes species to weakly react was Zymomonas mobilis.
DNA Fingerprinting
- RAPD-PCR
Random amplification of polymorphic DNA (RAPD-PCR) employs short primers (8-12 nucleotides long) with arbitrary sequences that bind nonspecifically to template bacterial DNA, which results in amplification of random, repetitive regions of template DNA, thereby providing a unique profile for bacterial identification. Rossetti et al. identify and classify over a thousand lactic acid bacteria (LAB) strains isolated mainly from dairy products, cheese in particular, through species-specific PCR and RAPD-PCR typing. The utilization of the M13 sequence as a primer yielded unique RAPD-PCR fingerprinting profiles, which were systematically analyzed using BioNumerics™ software. This analysis method arranged the strains into 11 major RAPD-PCR 'library units' which corresponded to specific LAB species including Lactobacillus casei/paracasei, Lactobacillus plantarum, Lactobacillus rhamnosus, and more. Further, the authors emphasized the potential of using their defined library to easily identify new bacterial isolates. This proposed method, they suggest, could enhance previously identified strains, minimize redundancy, and provide a technologically beneficial LAB strain collection. Furthermore, they propose that this approach can be applied to identify LAB strains isolated from other food ecosystems.
- RFLP
Restriction fragment length polymorphism (RFLP) is a method for identifying bacterial strains using unique fingerprints, which relies on the presence of variations in homologous DNA sequences. This PCR-based method employs restriction enzymes, which can recognize and cut amplified DNA (PCR product) into DNA fragments of different lengths. Electrophoresis of the enzyme digests distinguishes individuals, populations, or species or to pinpoint the locations of genes within a sequence. Laguerre et al.'s investigation encompassed forty-eight strains, representing a diverse spectrum including eight recognized Rhizobium species, two novel Phaseolus bean Rhizobium genomic species, Bradyrhizobium species, Agrobacterium species, and unclassified rhizobia derived from various host plants. These strains underwent examination via restriction fragment length polymorphism (RFLP) analysis targeting the 16S rRNA genes, amplified through PCR. Integration of data derived from RFLP analysis utilizing nine distinct endonucleases revealed the identification of twenty-one composite genotypes. Remarkably, species assignments were found to align entirely with established taxonomic classifications. Evaluation of genetic relationships, both inter- and intra-genera, based on these data exhibited robust concordance with previously published findings, particularly those derived from DNA-rRNA hybridizations and sequence analyses of 16S rRNA genes. Consequently, the PCR-RFLP methodology employed in this investigation affirms its efficacy as a rapid tool for the precise identification of root nodule isolates and the discrimination of novel taxa.
- AFLP
Amplified fragment length polymorphism (AFLP) employs restriction enzymes (usually a pair) to fragment genomic DNA and then amplifies a subset of restriction fragments using ligated adaptors. This amplification is achieved by using primers that are complementary to the adaptor sequences but also have certain unique nucleotides. Therefore, only a small number of restriction fragments are selectively amplified. AFLP fingerprints are then analyzed using gel electrophoresis, yielding a set of distinct DNA fragments from a single bacterial genomic DNA, which offers high specificity and discriminatory potential in the absence of any prior genome sequence knowledge.
Microarray
The DNA microarray, also known as a gene chip, is a well-established technique employed for measuring gene expression levels. The entire process unfolds on a specialized glass or silicon chip, where thousands of genes can be detected in a single reaction. This allows for a rapid and accurate high-throughput characterization of microbes.
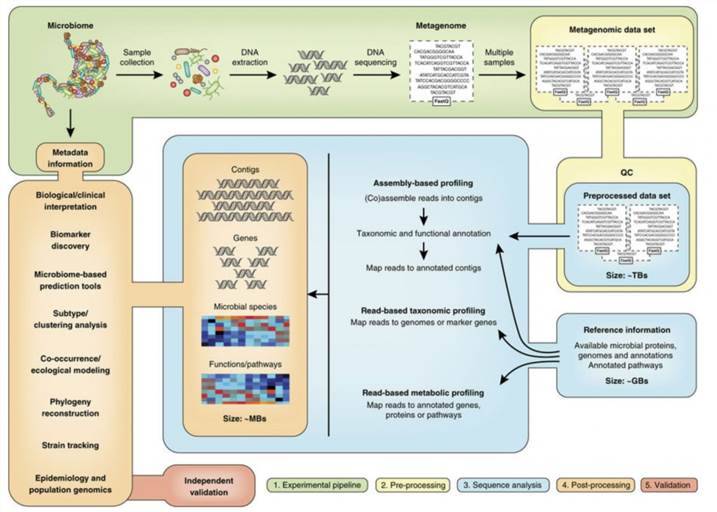
Metagenomics
Metagenomics is a culture-independent technology that can provide insights into the collective genome of a microbial community, indicating microbial diversity and ecology in specific environments. Metagenomics is generally based on next-generation sequencing, which reads billions of DNA base pairs in a single run. With abundant and comprehensive genetic information, microbial metagenomics provides a solid basis for the design and analysis of the omics study of microbiome characteristics. Abayasekara et al.'s study examine the challenges of diagnosing polymicrobial and fungal infections, noting that while molecular detection remains the premiere method for species identification in clinical samples, the limitations of Sanger sequencing present formidable barriers in the treatment of polymicrobial infections. They underscore the transformative role of Next Generation Sequencing (NGS) in the metagenomics field, yet lament its lack of commercial application in the detection of infectious diseases. In response to this, they introduce Credence Genomics' Rapid Infection Detection™ test. This test employs NGS technology in conjunction with molecular diagnostics to examine bacterial 16S rRNA genes and fungal ITS1 gene regions within a clinical sample, thereby facilitating a highly precise species identification. The authors compare the effectiveness of 16S and ITS1 metagenomic identification against conventional culture techniques for clinical samples, adopting the culture results as the gold standard for this comparison. The results reveal a concordance rate of 91.8% with culture-positive specimens and a 52.8% rate for culture-negative samples. Notably, they also detect fungal species in 10.3% of the specimens, which were not investigated by culture. Their study concludes that 16S-based metagenomic identification of bacterial species within a clinical specimen aligns with conventional culture-based techniques. They propose that, when combined with clinical information, this approach can significantly improve the precision and efficacy of infectious disease diagnosis tools.
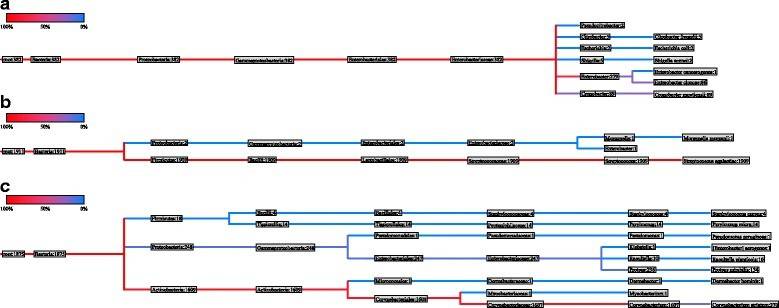 Figure 8. Phylogenetic tree of urine specimen (a) nasogastric aspiration specimen (b) and pus swab (c). (Abayasekara et al., 2017)
Figure 8. Phylogenetic tree of urine specimen (a) nasogastric aspiration specimen (b) and pus swab (c). (Abayasekara et al., 2017)
Examples of Microbial Detection and Identification Using Different Methods
Candida species constitute a major group of fungal pathogens in humans. Candidal infections rank as the fourth most common nosocomial infections globally. Given that Candida albicans is the most prevalent species causing candidal infections, accurate identification of Candida albicans from other species is crucial due to variations in antifungal susceptibility profiles. Ashraf et al. conducted a study to evaluate and compare several standard phenotypic and genotypic techniques for the identification of Candida albicans isolates obtained from 75 patients diagnosed with respiratory tract infections, urinary tract infections, or candidemia. The isolates underwent specific identification through germ tube testing, growth on chromogenic agar, and polymerase chain reaction (PCR). The methods employed and the results obtained are detailed below:
Germ tube method: Identification was conducted by observing the formation of germ tubes in human serum after incubation.
Subculture on Candida chromogenic agar medium: Identification was based on colony color observed on Candida chromogenic agar plates.
Polymerase chain reaction (PCR): Identification was performed by detecting specific C. albicans DNA sequences.
Table 1. Identification of different Candida species on chromogenic agar
| Yeast species | Colony color |
|---|---|
| C.albicans | Green |
| C.krusei | Pink, fuzzy |
| C.tropicalis | Dark blue |
| Other Candida species | Beige-yellow creamy color |
Based on the utilization of different methods, the identification outcomes for C. albicans are delineated as follows:
Germ tube method: Among the isolates tested, 48 demonstrated positive germ tube formation, while 27 were negative, resulting in an identification accuracy of 84.9%.
Subculture on Candida chromogenic agar medium: Notably, 51 isolates exhibited green colonies, 6 formed pink colonies, 1 formed blue colonies, and 15 formed white creamy colonies, with an identification accuracy of 96.3%.
PCR: Remarkably, all 53 isolates were successfully identified as C. albicans through PCR, boasting a noteworthy identification accuracy of 100%.
In essence, PCR showcased exceptional sensitivity, achieving a flawless 100% identification rate for C. albicans, thereby surpassing the efficacy of the other two methods. While Candida chromogenic agar medium demonstrated commendable accuracy, it exhibited limitations in distinguishing between various Candida species. Despite being regarded as the gold standard, the germ tube method displayed certain constraints within the context of this study.
Zakrzewski et al. investigates the identification of Enterobacterales strains isolated from fresh fish and shrimps, aiming to compare the effectiveness of different identification methods. The methods employed in the study include biochemical assays, sequencing, and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Biochemical identification was performed using the EnteroTest 24N set, while MALDI-TOF MS analysis was conducted using VITEK® MS. The results were compared with those obtained from 16S rRNA sequencing.
EnteroTest 24N involved fermentation and other biochemical assays, while MALDI-TOF MS utilized mass spectrometry for identification. Sequencing was performed on extracted DNA using 16S rRNA sequences. The results revealed discrepancies among the identification methods, with MALDI-TOF MS showing higher accuracy compared to EnteroTest 24N. MALDI-TOF MS correctly identified strains to the species level in 70.97% of cases, while EnteroTest 24N achieved only 50.0% accuracy. Additionally, MALDI-TOF MS demonstrated superior genus identification (90.32%) compared to EnteroTest 24N (approximately 70.0%).
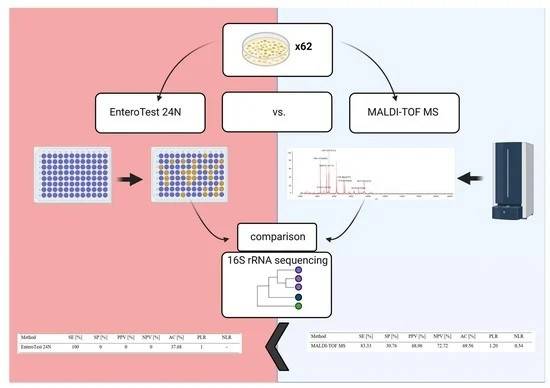 Figure 9. A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns. (Zakrzewski et al., 2022)
Figure 9. A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns. (Zakrzewski et al., 2022)
Overall, the study highlights the limitations of conventional identification methods and emphasizes the importance of confirming results using alternative techniques, particularly sequencing. The findings underscore the potential of MALDI-TOF MS as a rapid and accurate method for identifying Enterobacterales strains isolated from food samples.
Comparison of the Advantages and Disadvantages of Different Microbial Detection and Identification Methods
Comparing microbial detection and identification methods is crucial in biology, as it offers solutions for accuracy, reliability, sensitivity, specificity, cost-effectiveness, ease of operation, and applicability across various sample types and microbial targets. Differences in accuracy and reliability between methods help determine which is more reliable for specific microbes or sample types. Variation in sensitivity and specificity among microbial detection methods allows selection based on sample type or detection target. Assessing cost-effectiveness aids in choosing economical yet effective methods. Comparing ease of operation assists in selecting methods suitable for laboratory conditions and personnel skills. Variability in applicability across different microbial types or sample types helps identify the most suitable method for specific research or application areas.
Table 2. Advantages and Disadvantages of Different Microbial Detection and Identification Methods
| Method | Advantages | Disadvantages |
|---|---|---|
| Culture Methods |
|
|
| ELISA |
|
|
| MALDI-TOF MS |
|
|
| HPLC |
|
|
| Whole-Genome Sequencing |
|
|
| Ribotyping |
|
|
| Real-Time PCR |
|
|
| RAPD-PCR |
|
|
| RFLP |
|
|
| AFLP |
|
|
| Microarray |
|
|
| Metagenomics |
|
|
Harnessing a combination of diverse microbial detection and identification methods can compensate for limitations inherent to each individual method, bolstering accuracy and reliability of results. For instance, coupling conventional cultivation techniques with molecular biology technology allows for rapid identification and quantitative analysis of specific microorganisms. Another prime example is the integration of gene sequencing technology with spectrometry, enabling precision in microbial identification and genome analysis. The comprehensive application of multiple methods empowers researchers to attain an all-encompassing understanding of microbial community structure and functions. This provides robust data support for research and practical applications in related fields. For instance, in environmental microbiology, the blend of various detection methods bolsters the precision of assessing levels of microbial contamination in the environment, as well as monitoring ecosystem health. In the field of clinical microbiology, leveraging different detection methods enhances both the timeliness and diagnostic ability for pathogenic microorganisms. This, in turn, assists in the formulation of more effective treatment schemes and control measures.
References
- Ferone M, Gowen A, Fanning S, et al. Microbial detection and identification methods: Bench top assays to omics approaches. Comprehensive Reviews in Food Science and Food Safety, 2020, 19(6): 3106-3129.
- Anyansi C, Straub T J, Manson A L, et al. Computational methods for strain-level microbial detection in colony and metagenome sequencing data. Frontiers in Microbiology, 2020, 11: 538426.
- Nemati M, Hamidi A, Dizaj S M, et al. An overview on novel microbial determination methods in pharmaceutical and food quality control. Advanced pharmaceutical bulletin, 2016, 6(3): 301.
- Diba K, Kordbacheh P, Mirhendi S H, et al. Identification of Aspergillus species using morphological characteristics. Pakistan journal of medical sciences, 2007, 23(6): 867.
- Lee H J, Suh J T, Kim Y S, et al. Typing and antimicrobial susceptibilities of methicillin resistant Staphylococcus aureus (MRSA) strains isolated in a hospital in Korea. Journal of Korean Medical Science, 2001, 16(4): 381.
- Cherkaoui A, Schrenzel J. Total laboratory automation for rapid detection and identification of microorganisms and their antimicrobial resistance profiles. Frontiers in Cellular and Infection Microbiology, 2022, 12: 807668.
- Zhao Q, Lu D, Zhang G, et al. Recent improvements in enzyme-linked immunosorbent assays based on nanomaterials. Talanta, 2021, 223: 121722.
- Ashfaq M Y, Da'na D A, Al-Ghouti M A. Application of MALDI-TOF MS for identification of environmental bacteria: A review. Journal of environmental management, 2022, 305: 114359.
- Abayasekara L M, Perera J, Chandrasekharan V, et al. Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: a comparative study. BMC infectious diseases, 2017, 17: 1-11.
- Hasman H, Saputra D, Sicheritz-Ponten T, et al. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. Journal of clinical microbiology, 2014, 52(1): 139-146.
- Salloum G, Meugnier H, Reyrolle M, et al. Identification of Legionella species by ribotyping and other molecular methods. Research in microbiology, 2002, 153(10): 679-686.
- Haakensen M, Dobson C M, Deneer H, et al. Real-time PCR detection of bacteria belonging to the Firmicutes Phylum. International journal of food microbiology, 2008, 125(3): 236-241.
- Rossetti L, Giraffa G. Rapid identification of dairy lactic acid bacteria by M13-generated, RAPD-PCR fingerprint databases. Journal of Microbiological methods, 2005, 63(2): 135-144.
- Laguerre G, Allard M R, Revoy F, et al. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Applied and environmental microbiology, 1994, 60(1): 56-63.
- Ashraf A. Kadry, Amira M. El-Ganiny, Ahmed M. El-Baz. Comparison of methods used in identification of Candida albicans. Research J. Pharm. and Tech. 2018; 11(3): 1164-1168.
- Zakrzewski A J, Zarzecka U, Chajęcka-Wierzchowska W, et al. A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns. Pathogens, 2022, 11(4): 410.