Genomics has changed how we understand biology. But often, the biggest challenge in cutting-edge research is simply not having enough high-quality DNA. Whether it’s from a single cell, tiny bits of DNA in blood (like from tumors), ancient specimens, or even crime scene evidence, getting enough genetic material without mistakes or missing pieces is critical. This is where Whole-Genome Amplification (WGA) becomes incredibly important. WGA is a set of advanced lab methods designed to make many copies of an entire genome, even when you start with very little. This article provides a comprehensive overview of whole-genome amplification (WGA) technologies, tracing their evolution from PCR-based methods to advanced isothermal and microfluidic systems, and highlights their transformative impact across fields such as single-cell genomics, cancer diagnostics, reproductive medicine, forensics, and ancient DNA research.
WGA’s Core Role in Modern Genetics
WGA first appeared in 1992 and was a huge leap forward from standard PCR, which only copies specific DNA sections. WGA’s main goal is clear: to create lots of DNA copies that perfectly match the original genome. This lets scientists perform detailed genetic analyses that were impossible before due to limited sample size. WGA isn’t just a small improvement; it’s a key tool for fields like forensics, genetic disease research, next-generation sequencing (NGS), and the rapidly growing area of single-cell genomics. Being able to turn tiny amounts of starting DNA (picograms) into much larger amounts (micrograms) solves basic quantity and quality problems in many biological studies.
How WGA Works: Old Ways vs. New Ways of Copying DNA
WGA technology has come a long way, like going from an old-fashioned copy machine to a super-fast, smart printer. There are two main types of WGA methods: those that use PCR (which involves heating and cooling) and those that work at a steady temperature (isothermal). Each has its own pros and cons as scientists keep working to make WGA better at copying DNA completely, evenly, and accurately.
Early Methods: Using PCR to Copy DNA
The first WGA methods adapted the PCR process to copy entire genomes.
1. PEP-PCR (Primer Extension Preamplification)
This was one of the earliest methods. It uses random primers (short DNA pieces that can attach almost anywhere). The copying process involves cycles of lower-temperature attachment, then gradually warming up to a higher copying temperature. This helps the primers attach broadly across the DNA. While it could copy a lot of DNA from a single cell, its random nature and less strict settings often led to uneven copying, making it hard to get precise measurements.
2. DOP-PCR (Degenerate Oligonucleotide Primed PCR)
This method improved on PEP-PCR by using primers with fixed ends and a random middle. It starts with a few low-temperature cycles for broad attachment, then switches to higher temperatures for fast copying. DOP-PCR works for small samples, making DNA pieces around 500 building blocks long. But because it copies so quickly (exponentially), any small unevenness at the start gets magnified greatly, so the overall DNA coverage isn’t always ideal.
3. T-PCR (Tagged Random Primer PCR)
This method adds a fixed "tag" to the random primers. This tag helps make the second round of copying more precise. It’s efficient, but the process can be tricky, and you might lose some precious original DNA during cleanup.
4. LM-PCR (Ligation-Mediated PCR)
This technique first breaks the original DNA into pieces. Then, special adapters are attached to the ends of these pieces. Finally, PCR uses primers that match these adapters to copy the broken-up DNA. LM-PCR is very sensitive and copies well, but it involves many steps, making it complex.
5. LA-PCR (Long and Accurate PCR) and IRS-PCR (Interspersed Repetitive Sequence PCR)
LA-PCR helps copy longer DNA pieces more accurately by mixing different types of DNA copying enzymes. IRS-PCR uses natural repeating DNA sections as starting points for copying. However, because these repeats aren’t always spread evenly, the copying can be quite uneven, limiting its general use.
Overall, while these PCR-based WGA methods were important first steps, they struggled with common issues: they could copy some parts of the DNA more than others and sometimes introduced mistakes into the copies.

Figure 1. PCR-based technologies of whole-genome amplification methods.( Van Loo, P. et al, 2013)
The New Frontier: Constant-Temperature WGA
Constant-temperature (isothermal) WGA methods were a big step forward. They do all the copying at a steady temperature, which puts less stress on the DNA and often leads to more accurate and efficient copying.
1. MDA (Multiple Displacement Amplification)
MDA is a cornerstone of constant-temperature WGA. It uses a special enzyme called phi29 DNA polymerase. This enzyme is great because it can copy new DNA strands while pushing aside existing ones, and it’s very good at making long DNA copies (tens to hundreds of thousands of building blocks long). It also has a very low error rate (it makes very few mistakes). MDA uses random primers that attach at many spots. The process is simple and gives lots of accurate DNA copies. However, with very tiny starting DNA amounts, it can still show random copying biases (unevenness) or miss some gene versions. Newer methods like digital droplet MDA (ddMDA) and emulsified MDA (eMDA) help fix this by dividing the reactions into millions of tiny droplets, making the copying more even.
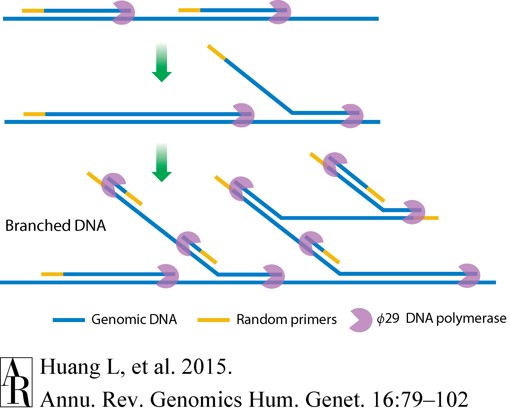
Figure 2. Schematic of multiple displacement amplification (MDA).( Huang, L. et al, 2015)
2. MALBAC (Multiple Annealing and Looping-Based Amplification Cycles)
This is one of the most advanced WGA techniques. It cleverly combines parts of MDA and PCR to minimize copying bias. It uses special primers, and early DNA copies are encouraged to form circular "loops." This "looping" prevents them from copying too fast, making the overall copying process more linear and much more even across the genome (up to 93% from a single cell). This makes MALBAC very useful for finding tiny changes in single cells.
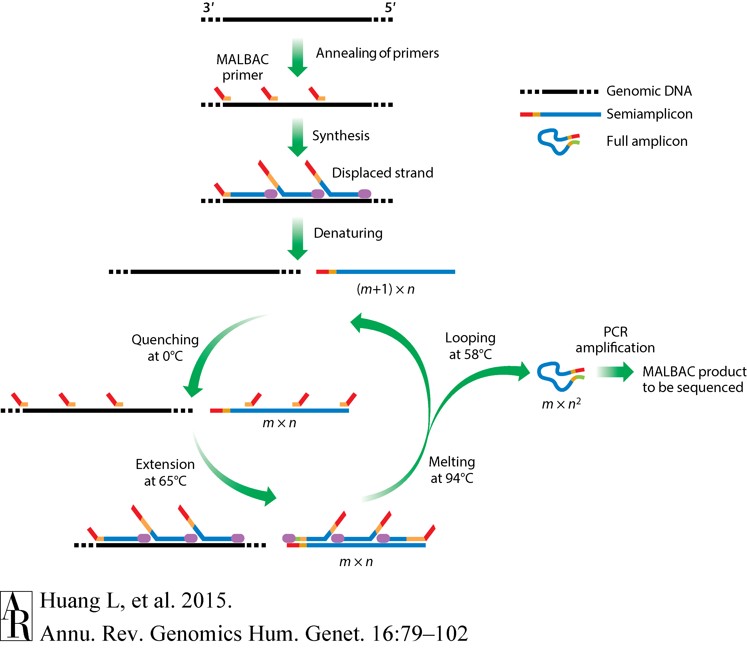
Figure 3. Schematic of multiple annealing and looping–based amplification cycles (MALBAC).( Huang, L. et al, 2015)
3. RCA (Rolling Circle Amplification)
RCA works with circular DNA. A short primer attaches to the circle, and a DNA copying enzyme continuously extends, making a long chain of repeated copies. RCA is good for specific tasks like copying circular DNA.
4. LIANTI (Linear Amplification via Transposon Insertion)
This is a very new WGA method for single cells that avoids fast, exponential copying. It uses a "jumping gene" enzyme called Tn5 transposase to insert special DNA pieces into the original DNA. These inserted pieces then cause many RNA copies to be made, which are then turned into DNA. By relying on this linear copying, LIANTI greatly reduces copying errors and biases, offering better accuracy for detecting changes in gene numbers and single-base changes. However, it’s a multi-step process, which can make it more complex.
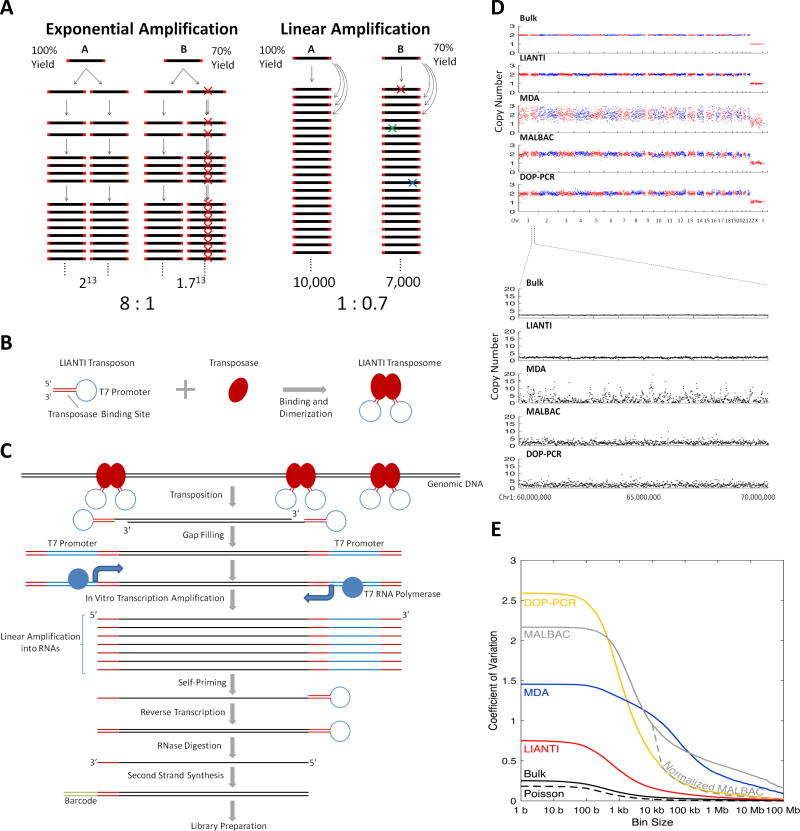
Figure 4. LIANTI single-cell whole genome amplification scheme.( Chen, C. et al, 2017)
The Power of Working Together: Microfluidics and WGA
Microfluidic technology has also had a big impact on WGA. Microfluidic chips are like tiny "factories" that can precisely control very small amounts of liquid. When combined with WGA, this means:
1) Tiny Samples: Works with extremely small amounts, even single cells.
2) Fast Reactions: Reactions happen quicker in tiny spaces.
3) High Sensitivity: It’s better at finding faint signals.
4) No Contamination: The sealed chips help prevent samples from mixing.
5) Reduced Bias: Microfluidic systems can put individual cells or DNA molecules into millions of tiny droplets. This helps ensure each tiny reaction starts with a similar amount of DNA, making the overall copying much more even and improving the detection of genetic changes.
While there are still some challenges with these tiny chips, combining microfluidics with WGA promises even more accurate and high-volume genetic analysis in the future.
WGA’s Impact: Changing Research and Medicine
WGA’s ability to turn tiny amounts of DNA into abundant data has made it an essential tool across many areas of biology and medicine, moving from basic research into real-world diagnostic uses.
Advancing Single-Cell Genomics
WGA is fundamental to single-cell genomic analysis, providing the necessary DNA for sequencing from individual cells. This capability is vital for:
1) Understanding Differences: It allows scientists to map the genetic makeup of individual cells within what might seem like uniform groups, revealing small but important variations in how tumors evolve, how brain cells differ, and how organisms develop. This level of detail is often impossible to get from analyzing large groups of cells together.
2) Solving Sample Limits: It overcomes the problem of having very little starting material, enabling genetic studies on rare cell types, old samples, or cells obtained through difficult biopsy procedures (like laser-guided cell removal or from circulating tumor cells in blood).
3) Improving Analysis Detail: Instead of simply averaging genetic signals from millions of cells, WGA lets us look at individual cells. This helps detect rare mutations and identify distinct groups of cells within a sample.
Changing Cancer Research and Personalized Treatment
In cancer research and patient care, WGA has become a game-changer:
1) Mapping Tumor Differences: Copying DNA from individual tumor cells allows for a very detailed look at how a tumor change over time, its specific mutations, and how it becomes resistant to treatments. This deep understanding helps guide more precise and tailored treatment plans.
2) Liquid Biopsy: Non-Invasive Cancer Monitoring: WGA makes it possible to amplify tiny amounts of circulating tumor DNA (ctDNA) or circulating tumor cells (CTCs) found in a patient’s blood. This offers a powerful way to non-invasively detect cancer, monitor how well treatment is working, and track tumor changes in real-time. This dynamic approach helps manage cancer more personally.
3) Detecting Gene Copy Changes (CNV): WGA products, especially from MDA and MALBAC, are crucial for tests like Comparative Genomic Hybridization (CGH) and NGS-based CNV detection. These tests help map changes where genes are copied too many or too few times in tumor cells, which are often key drivers of cancer growth and drug resistance.
Protecting Future Generations: Reproductive Health and Preimplantation Genetics
WGA significantly improves diagnostic options in reproductive medicine:
1) Prenatal Diagnosis: If there’s limited fetal DNA from tests like amniocentesis, WGA can amplify it. This makes genetic testing for inherited diseases more reliable, giving parents crucial information before birth.
2) Preimplantation Genetic Diagnosis (PGD) and Screening (PGS): A core part of assisted reproduction, WGA allows doctors to amplify DNA from just a single cell or a few cells taken from an early embryo. This enables comprehensive genetic screening for chromosome problems (PGS) or specific gene disorders (PGD) before the embryo is placed in the womb. This greatly improves the success rates of IVF and lowers the risk of passing on genetic diseases. Studies show that WGA-enhanced PGD (e.g., MDA+PCR) can lead to more accurate diagnoses and more healthy embryos available for transfer.
Extending Reach: Forensics and Ancient Discoveries
WGA’s ability to copy DNA from difficult samples has also proven vital in other fields:
1) Forensic Genetics: In crime investigations, evidence often contains only trace amounts of DNA, which might be degraded or from multiple people. WGA amplifies these tiny amounts, allowing for reliable DNA profiling (STR and SNP genotyping) for identification and kinship analysis, significantly improving success rates for challenging cases.
2) Paleontology and Anthropology: WGA helps recover and amplify highly degraded DNA from ancient remains (like bones or preserved tissues). This opens doors to studying evolutionary history, ancient populations, and ancient diseases.
Pushing Boundaries in Microbiology and Brain Science
WGA also plays a crucial role in expanding our knowledge of harder-to-access biological systems:
1) Microbial Genomics: It allows for the genetic study of microbes that can’t be grown in the lab or are very rare, directly from environmental or host samples. This vastly increases our understanding of microbial diversity, evolution, and their roles in ecosystems or disease.
2) Neuroscience: WGA provides the means to analyze the genetic differences within specific brain cell populations or even individual neurons. This sheds light on changes in DNA that might contribute to brain development, function, and disease.
What’s Next: Challenges and Big Hopes for WGA
WGA has come a long way, but there are still exciting goals:
1) Even Better Copies: Scientists are always working to make copying even more even, ensuring no part of the DNA is over or under-copied.
2) Perfect Accuracy: The goal is to make sure no "typos" or missing pieces are introduced during the copying process.
3) Easier to Use: Making WGA simpler and more automated will allow more labs and clinics to use it.
4) New Medical Tests: A promising area is making WGA even better for non-invasive tests from DNA floating in blood (like for cancer or fetal issues) and for studying modified DNA.
In short, WGA isn’t just about getting more DNA; it’s about getting high-quality, complete information. Its ongoing development means it will keep pushing the boundaries of what we can learn from even the tiniest samples, helping us understand life better and find new ways to diagnose and treat diseases.
Learn More
References
- Van Loo, P., & Voet, T. (2014). Single cell analysis of cancer genomes. Current opinion in genetics & development, 24, 82–91. https://doi.org/10.1016/j.gde.2013.12.004
- Huang, L., Ma, F., Chapman, A., Lu, S., & Xie, X. S. (2015). Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annual review of genomics and human genetics, 16, 79–102. https://doi.org/10.1146/annurev-genom-090413-025352
- Chen, C., Xing, D., Tan, L., Li, H., Zhou, G., Huang, L., & Xie, X. S. (2017). Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science (New York, N.Y.), 356(6334), 189–194. https://doi.org/10.1126/science.aak9787


 Sample Submission Guidelines
Sample Submission Guidelines