In biology science, transcription is the core biological process of transforming DNA genetic information into RNA and is the key link of gene expression regulation. Although both eukaryotes and prokaryotes rely on transcription to realize the transmission and expression of genetic information, due to the significant differences in cell structure, metabolic needs, and survival strategies, their transcription processes also show completely different characteristics. In-depth comparison of the transcription process between eukaryotes and prokaryotes not only helps to understand the basic laws of gene expression regulation, but also provides an important theoretical basis for exploring the evolution of life, analyzing the mechanism of disease occurrence, and developing new biotechnology.
This paper compares the differences between eukaryotes and prokaryotes in the transcription process, analyzes the causes and evolutionary significance, and introduces the application of related sequencing technology.
Overview of Eukaryotic and Prokaryotic Transcription
Transcription is the key process of transferring genetic information from DNA to RNA, and it plays an important role in the initiation of gene expression in both eukaryotes and prokaryotes. However, due to the different cell structures and functions, there are many differences in the transcription process.
Prokaryotes Transcription
Prokaryotes, such as bacteria, have a simple cell structure, no nucleus covered by a nuclear membrane, and the genetic material, DNA, is concentrated in the quasi-nuclear region.
- The transcription process of prokaryotes is closely coupled with the translation process. That is, the translation process has already begun before the transcription is completed. This close relationship enables prokaryotes to quickly respond to environmental changes and synthesize the required protein.
- The core of prokaryotic transcription is the interaction between RNA polymerase and DNA template, which starts the transcription process by identifying specific promoter regions and generates RNA molecules such as mRNA.
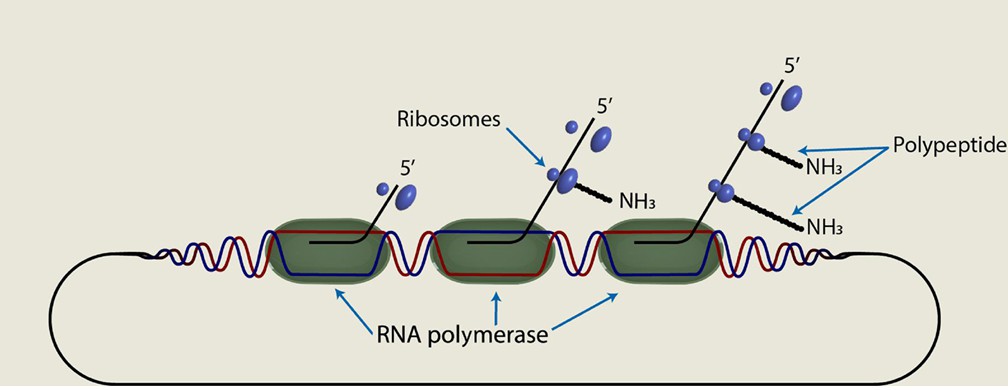
The coupled prokaryotic system of transcription and translation (Bell et al., 2020)
Eukaryotic Transcription
Eukaryotic cells are complex in structure, with a nuclear envelope. DNA mainly exists in the nucleus, and mitochondria and chloroplasts also contain a small amount of DNA.
- The transcription process of eukaryotes occurs in the nucleus, and the RNA produced by transcription needs a series of complex processes before it can be transported to the cytoplasm for translation.
- The transcription regulation mechanism of eukaryotes is more elaborate and complex, involving the synergistic effect of various transcription factors and regulatory elements to ensure the accurate expression of genes under specific time, space, and physiological conditions.
What is Differences in Eukaryotic and Prokaryotic Transcription
Transcription, the key link of genetic information transmission, shows significant differences between eukaryotes and prokaryotes. Eukaryotic cells have a complex structure, a fine transcription process, and multi-layer regulation; prokaryotic cells are simple in structure, and transcription pays more attention to a rapid response environment. These differences run through the initiation, regulation, termination, and post-processing of transcription, which deeply affect the gene expression and life activities of the two kinds of organisms.
Promoter Structure Difference
A. Prokaryotes promoter
a). The promoter structure of prokaryotes is relatively simple, which usually contains two conserved sequences -10 region (Pribnow box) and the -35 region. The common sequence of the -10 region is TATAAT, and the common sequence of the -35 region is TTGACA. These two regions are the key sites for RNA polymerase recognition and binding.
b). The σ factor in RNA polymerase holoenzyme (α β β’ ω σ) can specifically recognize the -10 region and -35 region of the promoter, and guide RNA polymerase to bind to the promoter to form a closed transcription initiation complex. Subsequently, the double-stranded DNA was partially untied to form an open transcription initiation complex, and the transcription process was officially started.
B. Eukaryotes promoter
a). The promoter structure of eukaryotes is more complex and diverse, and the core promoter region contains many elements, such as the TATA box, initiator (Inr), downstream promoter element (DPE), and so on.
b). TATA box is usually located about 25-35 base pairs upstream of the transcription initiation site, and its common sequence is TATAAA, which is similar to the -10 region of prokaryotes and is an important regulatory element for transcription initiation.
c). There are a large number of cis-acting elements in eukaryotes, such as enhancers and silencers, which can be located upstream or downstream of the transcription initiation site or inside the gene, and can remotely regulate the transcription initiation by combining with transcription factors.
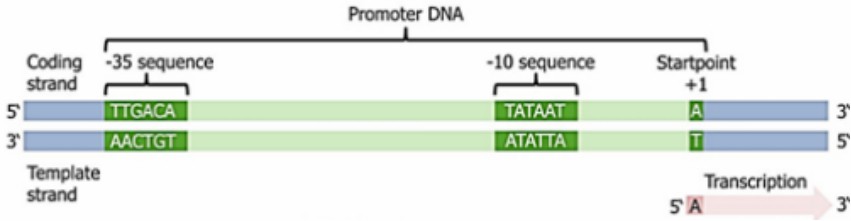
Promoter sequence in the prokaryotic cell (Prakash et al., 2023)
RNA Polymerase Difference
Prokaryotes have only one RNA polymerase, which is responsible for synthesizing all types of RNA, including mRNA, tRNA, and rRNA. The RNA polymerase is composed of several subunits, in which α, β, β’ ω are responsible for catalyzing RNA synthesis, and the σ factor is responsible for recognizing promoters. Different σ factors can recognize different promoter sequences, thus regulating the expression of different genes, so that prokaryotes can quickly adapt to environmental changes.
Eukaryotes contain three different RNA polymerases, namely RNA polymerase I, RNA polymerase II, and RNA polymerase III, which are responsible for transcription of different types of RNA.
- RNA polymerase I is mainly responsible for transcription of rRNA gene and synthesis of ribosomal RNA precursor.
- RNA polymerase II is responsible for transcription of protein coding genes, generation of mRNA precursors, and some noncoding RNA.
- RNA polymerase III is mainly responsible for transcription of tRNA gene, 5S rRNA gene and other small non-coding rnas.
Each RNA polymerase has unique subunit composition and functional characteristics, and needs the assistance of different transcription factors in the process of transcription initiation.
Transcription Speed
The transcription speed of prokaryotes is relatively fast, and usually 40-100 nucleotides can be synthesized per minute. This is mainly because the prokaryotic cell structure is simple, transcription and translation are coupled, and there is no complicated nuclear membrane barrier and post-transcription processing, so that RNA polymerase can move quickly along the DNA template and synthesize RNA efficiently. This rapid transcription is helpful for prokaryotes to quickly synthesize the required protein when the environment changes, and to maintain the survival and growth of cells.
The transcription speed of eukaryotes is relatively slow, and about 20-40 nucleotides are synthesized per minute. This is because the transcription process of eukaryotes is limited by many factors.
- Eukaryotic DNA combines with histone to form a chromatin structure, which will hinder the movement of RNA polymerase. It is necessary to untie the combination of DNA and histone through chromatin remodeling and create conditions for transcription.
- The transcription initiation of eukaryotes needs the synergistic effect of various transcription factors and regulatory elements to form a complex transcription initiation complex, which is relatively time-consuming. In addition, RNA produced by eukaryotic transcription needs to go through a series of complex processing and modification processes, such as capping, tail adding, and splicing, which will also affect the overall speed of transcription.
The Arabidopsis complement of transcription factors (Riechmann et al., 2002)
Termination Signal Difference
Transcription termination signals in prokaryotes can be divided into ρ factor-dependent termination signals and ρ factor-independent termination signals.
- The termination signal, independent of the ρ factor, usually contains a GC-rich reverse repeat sequence followed by a poly-U sequence. When RNA polymerase is transcribed to the termination signal region, GC-rich reverse repeats will form a stem-loop, which will hinder the progress of RNA polymerase, and then the base pairing between poly-U sequence and poly-A sequence in the DNA template is weak, which makes RNA-DNA hybrid easy to dissociate, thus leading to transcription termination.
- The termination signal, dependent on the ρ factor, has no obvious special sequence characteristics. The ρ factor is a kind of protein with RNA-DNA helicase activity, which can bind to the newly born RNA chain and move to the 3′ end along the RNA chain. When encountering the stalled RNA polymerase, the ρ factor dissociates the RNA-DNA hybrid through its helicase activity and terminates transcription.
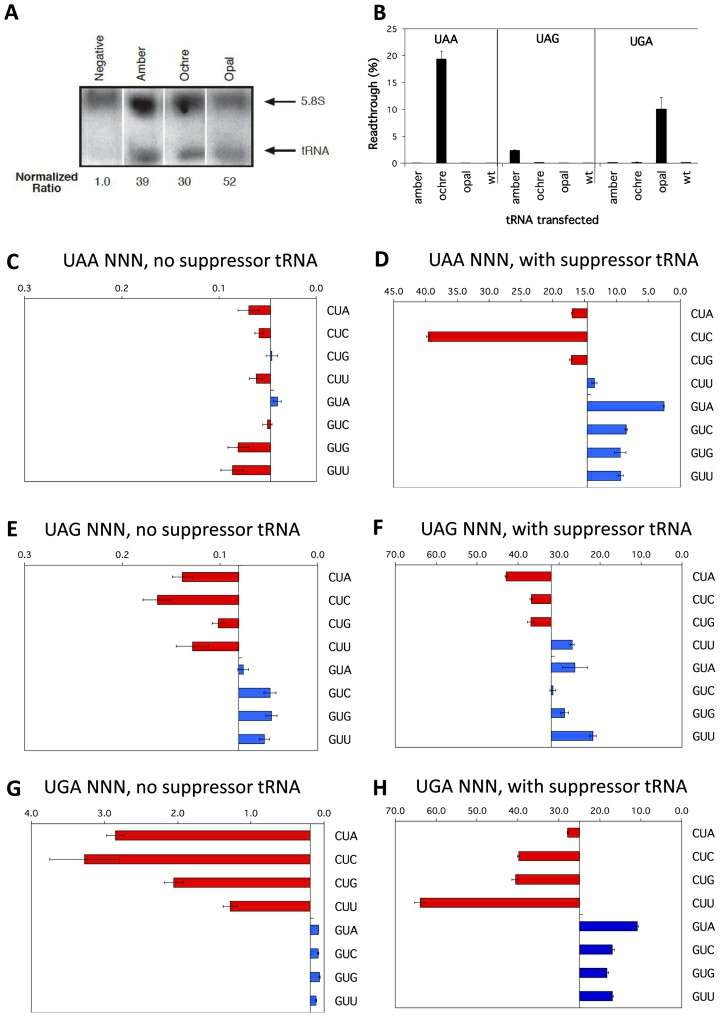
Competition by cognate tRNAs at termination signals (Cridge et al., 2018)
The transcription termination mechanism of eukaryotes is more complicated, and there are many studies on the transcription termination mechanism of RNA polymerase II at present. Transcription termination of RNA polymerase II is usually closely related to the 3′ end processing of mRNA. During the transcription process, when RNA polymerase II is transcribed to a specific termination signal region, it will trigger the 3′-end processing of mRNA, including cutting and tailing.
This process involves the interaction between various protein and nucleic acid elements. After cleavage, RNA polymerase II continues to transcribe for a certain distance, and then dissociates from the DNA template to complete transcription termination. Although the specific sequence characteristics of eukaryotic transcription termination signals are not completely clear, studies have shown that some cis-acting elements and transcription factors are involved in the regulation of transcription termination.
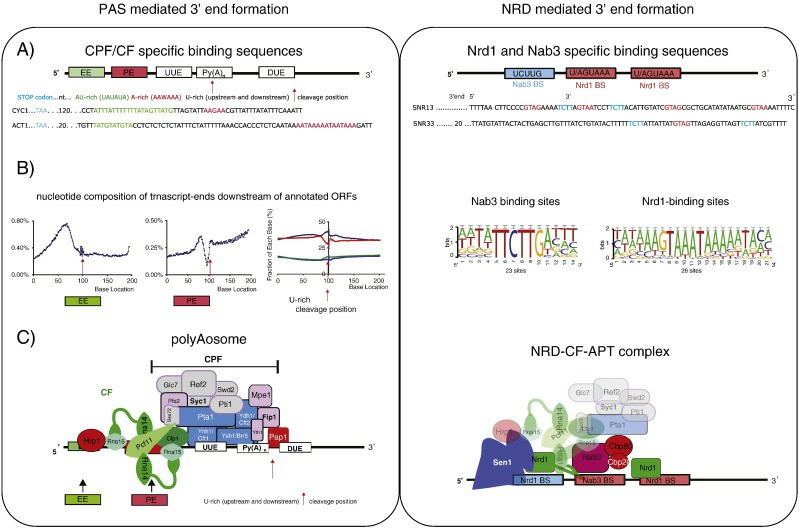
3′ end formation signals in yeast (Mischo et al., 2013)
Difference of Sequencing Technology
In the study of eukaryotic transcription, many sequencing technologies could be considered.
- RNA-seq is a commonly used technology that can comprehensively analyze gene expression, alternative splicing, and discover new transcripts, and help reveal gene expression differences and regulatory mechanisms.
- CAGE-seq is used to accurately determine the transcription initiation site, which is helpful to study the promoter structure and transcription initiation regulation.
- Long-read Sequencing can analyze complex alternative splicing isomers and complete transcript structures.
- scRNA-seq can analyze transcriptome at single cell level and reveal cell heterogeneity, which is of great significance to development and tumor biology research.
In the study of prokaryotic transcription, RNA-seq can analyze the changes in gene expression, clarify the boundary and abundance of transcripts, and explore the expression pattern of genes in the process of environmental response.
GRO-seq, PRO-seq, NET-seq and other technologies can monitor the transcription process in real time, accurately determine the dynamic changes of transcription initiation site and extension, and have unique advantages in studying the initiation and termination of prokaryotic transcription and the interaction between transcription factors and DNA.
Causes of Differences and Their Evolutionary Significance
It is no accident that there are significant differences between eukaryotes and prokaryotes in the transcription process. The differentiation of cell structure and the evolution of functional requirements are the root causes of differences: the simple cell structure of prokaryotes promotes efficient transcription mode, while the complex cell structure of eukaryotes shapes a fine regulation system. These differences have far-reaching significance in the evolution of life and promote the development of biodiversity and adaptability.
Causes of Differences
The fundamental reason for the difference in the transcription process between eukaryotes and prokaryotes lies in the difference in cell structure and function. Prokaryotes have a simple cell structure, lack of nuclear membrane-coated nucleus, and the coupling of transcription and translation. This structural feature makes prokaryotes need a fast and efficient transcription mechanism to adapt to the rapid changes in the external environment. Therefore, the transcription process of prokaryotes has the characteristics of simple promoter structure, a single RNA polymerase, fast transcription speed, and relatively direct transcription regulation.
Eukaryotic cells are complex in structure, with nuclei covered by a nuclear membrane. DNA and histone combine to form the chromatin structure, and gene expression regulation needs precise regulation at multiple levels. To ensure the accurate expression of genes under specific time, space, and physiological conditions, eukaryotes have evolved complex transcription initiation mechanisms, multi-level transcription regulatory networks, and elaborate post-transcription processing processes. These complex mechanisms enable eukaryotes to realize cell differentiation, development, and response to complex environmental signals.
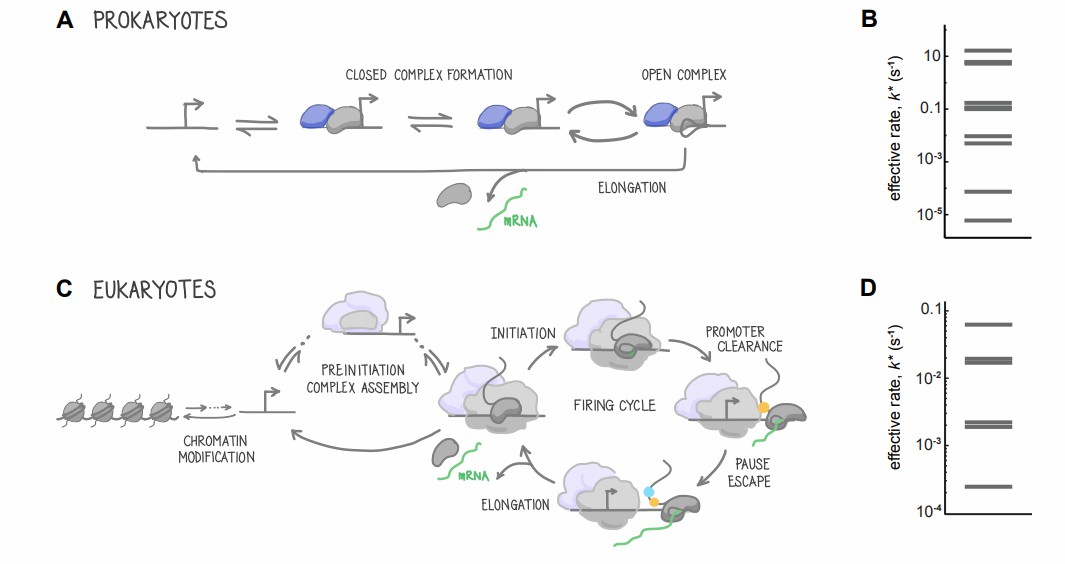
The transcription cycle in prokaryotes and eukaryotes (Clarissa et al., 2016)
Evolutionary Significance
The difference in the transcription process between eukaryotes and prokaryotes is of great evolutionary significance. For prokaryotes, the simple and efficient transcription mechanism enables them to rapidly synthesize the required protein in a constantly changing environment, so as to maintain the survival and reproduction of cells. Prokaryotes can occupy a place in the highly competitive ecological environment by quickly responding to environmental signals and regulating gene expression.
The complex and delicate transcription process of eukaryotes provides the basis for the evolution and diversity of organisms. The complex transcription regulation mechanism enables eukaryotes to differentiate cells and form tissues and organs, thus evolving various biological forms and functions. Post-transcriptional processing improves the stability of mRNA and the accuracy of translation, which is helpful to improve the efficiency and accuracy of gene expression. In addition, the transcription process of eukaryotes is coordinated with other biological processes (such as cell cycle regulation, signal transduction, etc.), forming a complex regulatory network, which enables eukaryotes to better adapt to the complex and changeable environment.
Conclusion
There are significant differences between eukaryotes and prokaryotes in transcription initiation, speed and regulation, termination signal and mechanism, post-transcription processing, and so on. These differences are determined by their different cell structures and functions, which have important biological significance and evolutionary value. The simple and efficient transcription mechanism of prokaryotes enables them to quickly adapt to environmental changes, while the complex and delicate transcription process of eukaryotes provides a guarantee for the evolution and diversity development of organisms.
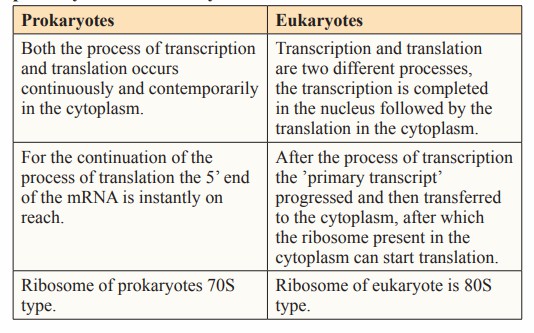
Difference between the process of the translation in prokaryotes and eukaryotes animals (Prakash et al., 2023)
In-depth study on the differences in the transcription process between eukaryotes and prokaryotes will help us better understand the basic laws of life activities, provide a theoretical basis for research and application in the field of biology, and also provide important clues for exploring the mysteries of biological evolution. With the deepening of research, we are expected to further reveal the regulation mechanism of the transcription process and its role in biological growth, development, and disease occurrence and development, and provide new ideas and methods for biotechnology, medicine, and other fields.
Learn more information, you can read:
- Sequencing Technology for Eukaryotic Transcription: An Exploration of Mechanisms, Technologies, Applications, and Challenges
- From Mechanism to Medicine: Understanding Eukaryotic Transcription Errors, Diseases and Treatments
- Eukaryotic Transcription: Key Stages, Regulatory Mechanisms, Products, and Significance
References
- Prakash D, Murali MS, et al. "Similarities and Differences of Expression of Gene in Prokaryotic and Eukaryotic Cells: A Critical Review." J Biotechnol Bioinforma Res. 2023 https://doi.org/10.47363/jlsrr%2F2023%281%29102
- Riechmann JL. "Transcriptional regulation: a genomic overview." Arabidopsis Book. 2002 1:e0085 https://doi.org/10.1199/tab.0085
- Cridge AG, Crowe-McAuliffe C., et al. "Eukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel." Nucleic Acids Res. 2018 46(4): 1927-1944 https://doi.org/10.1093/nar/gkx1315
- Mischo HE, Proudfoot NJ. "Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast." Biochim Biophys Acta. 2013 1829(1): 174-85 https://doi.org/10.1016/j.bbagrm.2012.10.003
- Clarissa S, Angela HD., et al. "Integrating regulatory information via combinatorial control of the transcription cycle." bioRxiv 2016 https://doi.org/10.1101/039339
- Bell PJL. "Evidence supporting a viral origin of the eukaryotic nucleus." Virus Res. 2020 289: 198168 https://doi.org/10.1016/j.virusres.2020.198168


 Sample Submission Guidelines
Sample Submission Guidelines