Non-coding RNA (ncRNA) is a type of RNA molecule that does not have the ability to encode proteins and plays a vital role in gene regulatory networks. In the past, people believed that the main function of RNA was to serve as a bridge for information transmission between DNA and protein, but with the deepening of research, the importance of non-coding RNA has gradually become prominent. Different types of non-coding RNA work synergistically in various physiological and pathological processes of cells to maintain the normal function of cells. This article will elaborate on the mechanism of action of three common ncRNAs .
What is Non-coding RNA
The essential difference between coding RNA and non-coding RNA lies in whether they have the ability to encode proteins. Coding RNA, such as messenger RNA (mRNA), carries genetic information and guides protein synthesis; non-coding RNA does not have this function. However, non-coding RNA occupies a core position in gene regulation. They can finely regulate gene expression at multiple levels, such as transcriptional level, post-transcriptional level, and translation level, affecting important biological processes such as cell proliferation, differentiation, and apoptosis. The discovery of non-coding RNA provides us with a new perspective for understanding gene regulatory networks and also brings new ideas for the diagnosis and treatment of diseases.
MicroRNA (miRNA) is a type of single-stranded non-coding RNA with a length of about 19-25 nt nucleotides, which mainly plays a role in the cytoplasm. Long non-coding RNA (lncRNA) is longer than 200 nucleotides, has a linear structure, and can be distributed in the nucleus and cytoplasm. CircRNA has a unique circular structure and is mainly present in the cytoplasm. The differences in length, structure and subcellular localization of these three types of non-coding RNA determine that they have different functions and mechanisms of action in gene regulation.
Table 1. Major ncRNAs
| type | length | structure | Subcellular localization |
|---|---|---|---|
| miRNA | 19 to 25 nt | Single Chain | Mainly in the cytoplasm |
| lncRNA | >200nt | Linear | Nucleus and cytoplasm |
| circRNA | Ring structure | ring | Mainly in the cytoplasm |
Mechanisms of miRNA Regulation of Gene Expression
miRNA participates in post-transcriptional gene expression regulation in plants and animals and is highly conserved. Most miRNA genes exist in the genome in the form of single copies, multiple copies, or gene clusters. Each miRNA can have multiple target genes, and several miRNAs can also regulate the same gene. This complex regulatory network can regulate the expression of multiple genes through one miRNA, or finely regulate the expression of a gene through a combination of several miRNAs.
miRNA Production Process
The miRNA gene is transcribed by RNA polymerase in the cell nucleus to obtain pri-miRNA (primary miRNA), which is about 300~1000 bases in length. After one processing, pri-miRNA becomes pre-miRNA (precursor miRNA), i.e., microRNA precursor, which is about 70~90 bases in length. After processing, the export protein transports the pre-miRNA from the cell nucleus to the cytoplasm, and the pre-miRNA in the cytoplasm is further cut into double-stranded miRNA after being cleaved by Dicer enzyme. Finally, under the action of AGO protein, only one chain of the double-stranded miRNA is retained, producing a mature miRNA of about 19 ~25 nt in length .

Figure1. Classical pathway of miRNA production. (Baril, P et al . 20 14)
It is speculated that most of the miRNAs that have been identified are generated by a single-stranded RNA precursor with a hairpin structure of about 70 bases in size, which is processed by the Dicer enzyme. It has a 5′ phosphate group and a 3′ hydroxyl group, and is a small RNA fragment of about 21-25nt in size, which is located at the 3′ or 5′ end of the RNA precursor.
Function Mechanism of miRNA
As a key factor in post-transcriptional regulation, miRNA is like a "molecular switch" for gene expression, precisely regulating the expression level of genes. Its core mechanism for regulating gene expression is mainly based on complementary pairing with target mRNA, thereby affecting the stability and translation efficiency of mRNA.
mRNA degradation
When miRNA is fully complementary to the target mRNA, it triggers the degradation of the mRNA. miRNA forms an RNA-induced silencing complex (RISC) with AGO protein, which recognizes and binds to the target mRNA, and then the endonuclease cuts the mRNA into fragments, resulting in rapid degradation of the mRNA and reduced protein synthesis.
Translation inhibition
If miRNA and target mRNA are not completely complementary, gene expression is mainly regulated by translation inhibition. RISC binds to the 3′ untranslated region (UTR) of mRNA, hindering the binding or extension of ribosomes, thereby inhibiting the translation process of proteins. For example, PTEN is an important tumor suppressor gene that regulates key processes such as cell proliferation, apoptosis, invasion and metastasis by inhibiting the PI3K/AKT/mTOR signaling pathway. However, miR-21 binds to the 3’UTR of PTEN, inhibiting its translation or promoting its mRNA degradation, thereby significantly reducing the expression level of PTEN protein. This regulatory effect inhibits the activity of PTEN, which in turn leads to abnormal activation of the PI3K/AKT signaling pathway, promoting the proliferation and survival of tumor cells (Chawra, H et al. 2024) .
The Role of lncRNA in Chromatin Remodeling
lncRNA plays a direct and critical role in chromatin remodeling and transcriptional regulation, just like a "spatial organizer" in the gene expression regulatory network. It can interact with biological macromolecules such as proteins and DNA to form a complex regulatory network and precisely regulate gene expression. By binding to specific proteins, lncRNA can be recruited to specific genomic sites, thereby affecting the structure and function of chromatin. In addition, lncRNA can also directly interact with DNA to form RNA-DNA hybrids, further affecting the transcription process of genes.
Regulating chromatin structural state
On the one hand, lncRNA can directly act on chromatin, bind to DNA, form a DNA-lncRNA-DNA triple-stranded complex, and affect the binding of transcription factors and gene expression. For example, during the inactivation of the X chromosome, the Xist lncRNA covers the entire X chromosome with its unique secondary structure, resulting in transcriptional silencing. On the other hand, lncRNA can recruit epigenetic modification enzymes such as DNMTs and histone methyltransferases (such as PRC2) to specific gene loci to induce or maintain specific epigenetic marks. Studies have found that Xist directly interacts with PRC2 through its RepA repeat sequence, introduces H3K27me3 modification into the X chromosome, and achieves gene silencing.
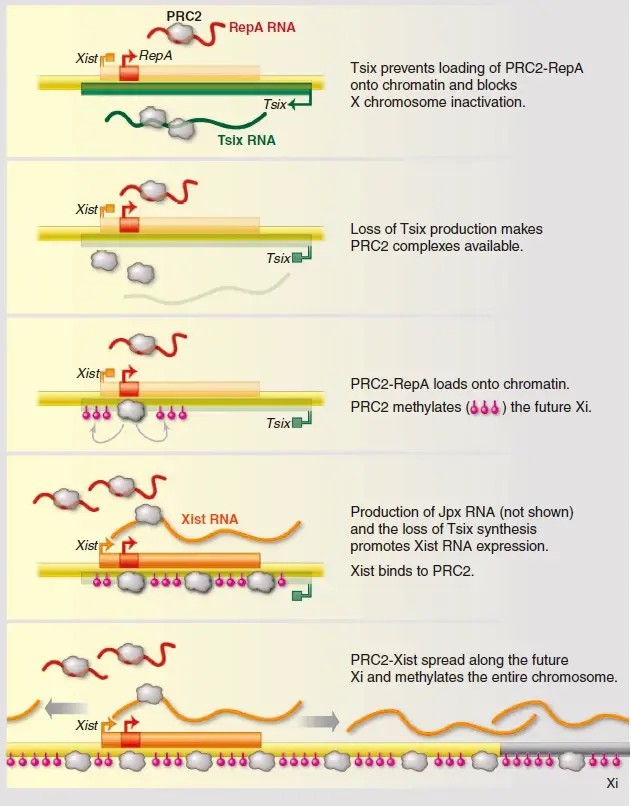
Figure 2. lncRNAs in X-chromosome inactivation. (Lee J. T 20 14)
Regulating DNA methylation
lncRNA also plays an important role in DNA methylation regulation. For example, lncRNAs such as Airn and H19 can bind to DNMTs and guide them to the promoter region of target genes, promote DNA methylation, and lead to gene silencing. This mechanism suggests that lncRNA can regulate DNA methylation levels through interaction with DNA methyltransferases (DNMTs), thereby affecting gene expression.
lncRNA can also affect DNA methylation through different mechanisms. For example, Xist lncRNA recruits polycomb repressive complex 2 (PRC2) to the X chromosome, methylating the CpG island promoter on the X chromosome, thereby inhibiting the expression of genes on the X chromosome. Demethylated lncRNAs, such as H19, can block DNA methylation modification mediated by DNA methyltransferase by inhibiting the supply of methyl groups for DNA methylation. This bidirectional regulatory mechanism shows that lncRNA is flexible and diverse in DNA methylation regulation .
Influence on chromatin remodeling and nucleosome positioning
lncRNA can also drive changes in chromatin structure and affect gene transcription activity by binding to chromatin remodeling complexes (such as SWI/SNF, ISWI, etc.). For example, lncRNA SchLAP1 and Myheart interact with the SWI/SNF complex, respectively, to affect gene expression. In addition, lncRNA Evf2 promotes the binding of the SWI/SNF complex to the Dlx5/Dlx6 gene enhancer, but inhibits its remodeling activity. This mechanism suggests that lncRNA can regulate chromatin structure and gene expression by interacting with chromatin remodeling complexes.
circRNA and Post-Transcriptional Regulation
circRNA is characterized by a covalently closed circular structure without open 5′ and 3′ ends. CircRNA is formed by precursor mRNA through an atypical splicing event, namely back-splicing, in which the downstream splicing site of the RNA molecule is connected to the upstream splicing site to produce a closed circular structure. CircRNA usually originates from the exon or intron region of the gene. They are stable in cells and can also play an important role in epigenetic modification.
Ring structure formation mechanism
The circular structure of circRNA is formed through the process of backsplice. In this process, first, the splicing sites at both ends of a specific exon in the precursor mRNA (pre-mRNA) are reversely connected. Then, the spliceosome recognizes these backsplice sites and connects the exons in reverse order to form a circular RNA molecule. Unlike linear RNA splicing, backsplice does not follow the traditional 5′ to 3′ splicing order, but crosses the normal splicing site, giving circRNA a unique structure and function.
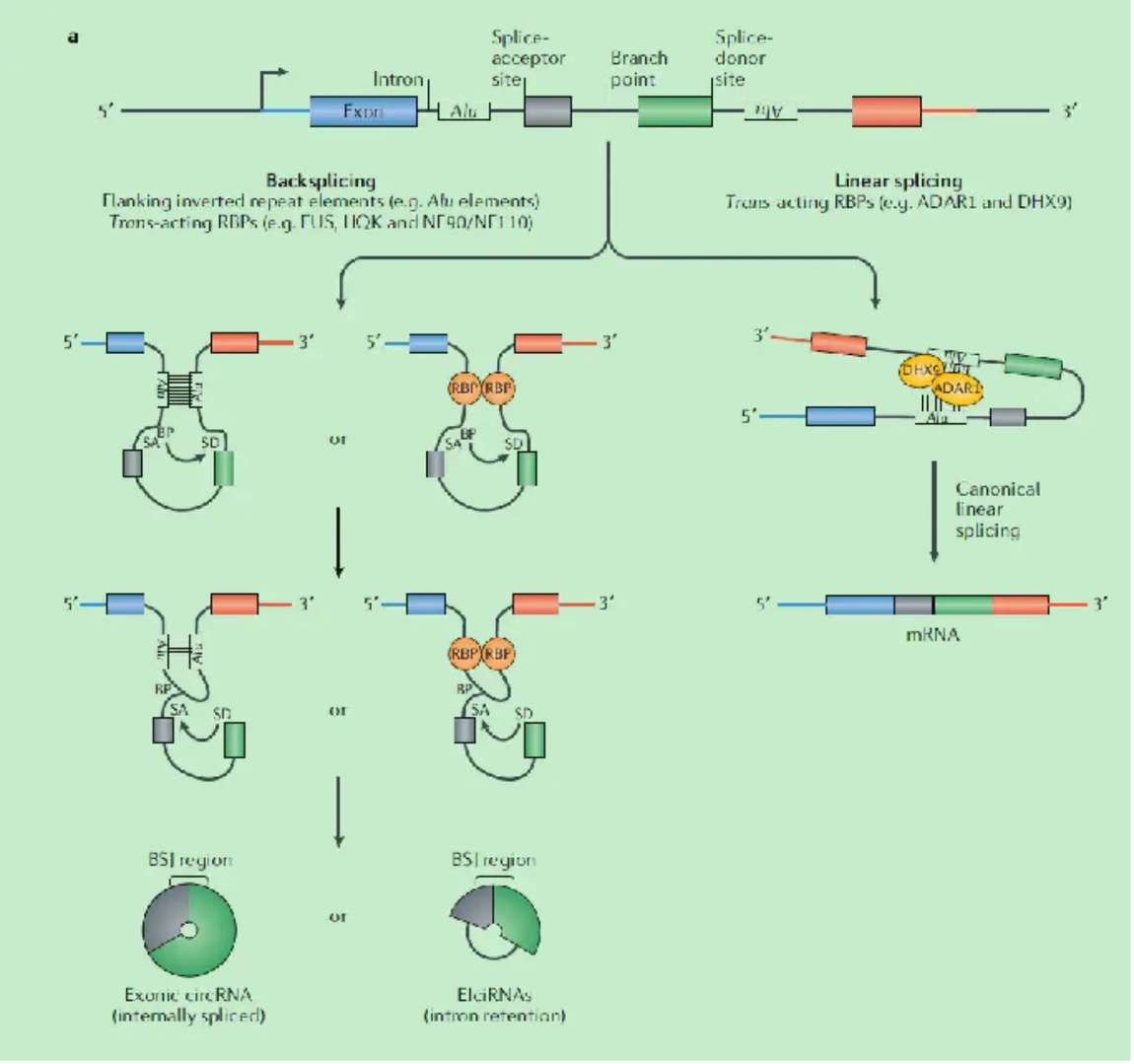
Figure 3.Biogenesis of circRNAs.(Kristensen, L. S, et al. 2019)
miRNA sponges: a regulatory model of molecular decoys
circRNA plays the role of miRNA sponge through miRNA response elements (MREs). circRNA contains multiple MREs complementary to miRNA, which can specifically adsorb miRNA. When circRNA binds to miRNA, miRNA cannot bind to target mRNA, thus relieving the inhibitory effect of miRNA on target mRNA. Taking ciRS-7 as an example, it contains a large number of binding sites complementary to miR-7 and can efficiently adsorb miR-7. In cells, ciRS-7 adsorbs miR-7, making it unable to inhibit the expression of its target mRNA, thereby affecting biological processes such as cell proliferation, differentiation and apoptosis.
Protein translation and interaction function
Small peptide translation: circRNA can mediate the translation of small peptides through the internal ribosome entry site (IRES). IRES can recruit ribosomes and initiate the translation process, allowing circRNA to encode small peptides. These small peptides have unique biological functions in cells and are involved in intracellular signal transduction and metabolic regulation.
Interaction with RBP: circRNA can interact with RNA-binding proteins (RBPs) and affect the function and localization of RBPs. This interaction can regulate processes such as gene expression, RNA metabolism, and intracellular signal transduction.
Research Techniques for Non-coding RNA
Non-coding RNA research technology continues to develop, from high-throughput global analysis to precise molecular mechanism analysis, providing powerful tools for in-depth understanding of the function and mechanism of non-coding RNA. Mainstream research technologies each have their own principles and application scenarios, and emerging technologies have also shown a good development trend.
RNA-seq: global expression profiling
RNA-seq is a powerful technology for global expression profiling. The first step is to build a library, extract RNA from the sample, reverse transcribe it into cDNA, and then perform operations such as fragmentation and adding adapters to construct a sequencing library. Then, sequencing is performed using a high-throughput sequencing platform to sequence the library and obtain a large amount of RNA sequence information. Finally, bioinformatics analysis is performed to screen out differentially expressed non-coding RNAs through comparison, quantification and other methods. In cancer research, RNA-seq has been successfully used to discover cancer-related non-coding RNA markers, providing an important basis for early diagnosis and treatment of cancer.
Single-cell transcriptome: unravelling heterogeneity
Single-cell transcriptome sequencing can reveal differences in the expression of non-coding RNA in different cell subpopulations. Its advantage is that it can overcome the limitations of traditional sequencing technology on the averaging of cell populations and accurately analyze the gene expression in each cell. In tumor microenvironment research, single-cell transcriptome sequencing can identify the expression characteristics of non-coding RNA in different cell subpopulations such as tumor cells and immune cells, which helps to gain a deeper understanding of the occurrence and development mechanism of tumors and provide accurate target information for personalized treatment.
CLIP-seq: Unraveling RNA-protein interactions
CLIP-seq: CLIP-seq (Cross-Linking and Immunoprecipitation followed by sequencing) is a cross-linking immunoprecipitation technique that fixes RNA and bound proteins together and then performs sequencing to analyze the interaction between RNA and protein. This technology can reveal RNA binding sites and regulatory mechanisms, providing a new tool for studying the regulatory network of non-coding RNA.
Conslusion
Non-coding RNAs play pivotal roles in gene regulation beyond the traditional central dogma. Through diverse mechanisms—such as mRNA degradation, chromatin remodeling, and miRNA sponging—these molecules influence essential biological processes like cell growth, differentiation, and apoptosis. Advances in sequencing and molecular biology tools continue to deepen our understanding of their regulatory networks, offering new insights into cellular function and disease pathogenesis, and paving the way for innovative diagnostic and therapeutic strategies.
References
- Baril, P., Ezzine, S., & Pichon, C. (2015). Monitoring the spatiotemporal activities of miRNAs in small animal models using molecular imaging modalities. International journal of molecular sciences , 16(3), 4947–4972. https://doi.org/10.3390/ijms16034947
- Chawra, HS, Agarwal, M.et al. (2024). MicroRNA-21’s role in PTEN suppression and PI3K/AKT activation: Implications for cancer biology. Pathology, research and practice , 254, 155091. https://doi.org/10.1016/j.prp.2024.155091
- Lee JT (2012). Epigenetic regulation by long noncoding RNAs. Science (New York, NY), 338(6113), 1435–1439. https://doi.org/10.1126/science.1231776
- Kristensen, LS, Andersen, MS, Stagsted, LVW, Ebbesen, KK, Hansen, TB, & Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nature reviews. Genetics , 20(11), 675–691. https://doi.org/10.1038/s41576-019-0158-7


 Sample Submission Guidelines
Sample Submission Guidelines