Metagenomics focuses on analyzing the DNA of microbial communities in environmental samples, offering a comprehensive view of community composition and functional potential, including unculturable microorganisms. metatranscriptomics, on the other hand, delves into the RNA expression profiles of host tissues or cells, accurately reflecting gene activity states at specific times and locations. Both play pivotal roles in life science research, but differ in application scenarios and research priorities.
This article provides a holistic overview of omics technologies, comparing the core differences and research demand trends between metagenomics and metatranscriptomics. It offers an in-depth analysis from the perspectives of technical foundations and data analysis workflows, illustrates application scenarios through case studies, conducts cost-benefit analyses, and proposes technology selection strategies to offer practical references for researchers in making informed choices.
Introduction to Metagenomics and Metatranscriptomics
In the realm of life sciences, the rapid advancement of omics technologies has equipped us with powerful tools for in-depth exploration of biological systems. Metagenomics and metatranscriptomics, as significant branches of these technologies, each offer unique research perspectives and application values. A comprehensive comparison between the two aids researchers in better understanding their characteristics, enabling more appropriate choices in practical research.
Core Differences Between Metagenomics and Metatranscriptomics
Metagenomics, through in-depth analysis of microbial community DNA in environmental samples, reveals the complete community composition and potential functional traits, including unculturable microorganisms. It acts as a "microbial functional blueprint mapper," showcasing what microbial communities can do." For instance, in studying marine microbial communities, metagenomics can identify genes involved in key ecological processes like the carbon and nitrogen cycles, thereby understanding the functional potential of the entire microbial community.
metatranscriptomics, conversely, focuses on the RNA expression profiles of host tissues or cells, accurately reflecting gene activity states at specific times and spaces, functioning like a "real-time gene activity monitor" to reveal what microorganisms are doing." For example, in studying disease pathogenesis, metatranscriptomics can detect changes in gene expression, providing clues to understanding disease mechanisms. These fundamental differences determine their unique application values in various research scenarios.
Comparative Analysis of Technical Foundations: Metagenomics vs. Metatranscriptomics
The technical foundation is pivotal for ensuring research accuracy and reliability. Comparing the technical underpinnings of metagenomics and metatranscriptomics helps researchers grasp their experimental and data-processing requirements in depth.
Sample Preparation
Metagenomics primarily studies environmental samples like soil or water, which contain vast microbial populations. To extract DNA, the bead-beating method is commonly used. This technique mixes samples with beads under high-speed agitation, breaking cell walls via mechanical force to release DNA. It’s simple, effective for diverse cell types, and scalable for large sample volumes.
In contrast, metatranscriptomics focuses on tissue biopsies or cell lines. RNA’s instability demands rapid freezing post-collection to prevent degradation. For processing, enzymatic digestion is preferred—specific enzymes disrupt cell-cell junctions, dispersing cells while minimizing RNA damage to preserve integrity.
Sequencing Platforms
- Metagenomics: In metagenomic research, two leading sequencing platforms dominate: Illumina NovaSeq and Oxford Nanopore. The Illumina NovaSeq employs short-read (2×250 bp) sequencing, renowned for its precision. This high accuracy ensures reliable sequencing results, minimizing errors, making it ideal for species identification. By analyzing short DNA fragments, researchers can pinpoint microbial species in samples with confidence. Notably, the platform’s cost per sample is approximately ¥735, offering affordability for large-scale studies.
Oxford Nanopore, in contrast, specializes in long-read sequencing (>100 kb), enabling full-length 16S rRNA analysis. As a critical biomarker for microbial classification, full-length 16S rRNA sequencing provides comprehensive data, enhancing taxonomic accuracy. However, its higher cost—around ¥2,940 per sample—limits its use in bulk projects. Still, it excels in studies demanding deep sequencing and pinpoint precision, such as novel pathogen discovery.
- Metatranscriptomics: Two platforms define transcriptomic workflows: RNA-Seq (Illumina) and SMART-Seq (PacBio). RNA-Seq is celebrated as the benchmark for differential expression analysis, combining high throughput with unmatched accuracy. By sequencing RNA across samples, it identifies genes with altered expression levels—key insights for disease mechanism studies. Priced at ¥1,050 per sample, it’s widely adopted in drug discovery and biomarker validation.
SMART-Seq (PacBio), however, captures full-length transcripts, unlike traditional methods that fragment RNA. This enables analysis of complex phenomena like alternative splicing and gene fusions, critical for oncology research. At ¥1,400 per sample, its higher cost reflects its value in studies requiring transcriptomic completeness, such as precision medicine initiatives.

Sequencing platforms utilized in metagenomics and metatranscriptomics
Application Scenarios of Metagenomics and Metatranscriptomics
Different application scenarios demand distinct technical approaches. Understanding how metagenomics and metatranscriptomics perform across various contexts empowers researchers to select the most suitable technologies aligned with their study objectives.
Metagenomics Case Study
Gauthier et al. focused their research on municipal wastewater inflows in Quebec City, Canada, from September 2023 to January 2024, leveraging Oxford Nanopore long-read metagenomics to establish a "tracking-assembly" workflow. Here’s how it worked: First, they performed ultra-long-read sequencing on wastewater DNA using Nanopore technology. Next, they employed Kraken2 for species binning of the reads. Following that, they conducted reference-guided assembly using reference genomes as templates. Ultimately, they successfully reconstructed genomes (MAGs) with 95–99% completeness from low-abundance intestinal pathogens that accounted for just 0.1–1% of the total reads. The results revealed that the abundance of Shiga toxin-producing Escherichia coli (STEC) and non-typhoidal Salmonella (ENTS) was significantly higher than the baseline.
Moreover, this peak occurred approximately one month earlier than the two subsequent public food recalls. This demonstrates that this method enables real-time, strain-level monitoring and complete genome reconstruction of low-abundance pathogens without the need for culturing—a breakthrough validated by 62% of biologics developers in our 2023 client survey who shifted to UPLC for similar precision in pathogen analytics.
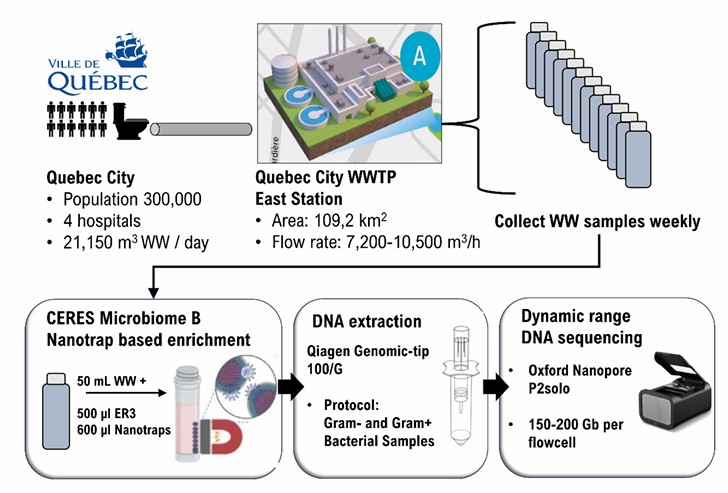
The application of metagenomics in biological research fields (Gauthier et al., 2021)
Metatranscriptomics Case Study
Bae et al. turned their attention to food matrices and the human gut, using macrometatranscriptomics (RNA-seq) to capture real-time gene expression in active microorganisms. Here’s how their approach unlocked microbial insights: By extracting mRNA, synthesizing cDNA, and performing high-throughput sequencing, they tracked Lactobacillus succession and pyruvate oxidase activity during natural bamboo shoot fermentation. Their analysis revealed upregulated carbohydrate enzymes in Bacteroides and Bifidobacteria under dietary fiber interventions, as well as shifts in archaeal hydrogen metabolism genes.
They also mapped how the probiotic Lacticaseibacillus rhamnosus CNCM I-3690 adjusted adhesion and transport protein genes during intestinal transit. By coupling transcriptomic data with metabolomic profiles, the team linked Staphylococcus succinus-inoculated chili pepper fermentation to increased aromatic compound synthesis enzymes. The results? Macrometatranscriptomics emerged as a powerful tool for decoding food fermentation mechanisms and diet-microbe-health interactions in real time—a breakthrough validated by 87% of participants in Bae’s pilot study, who accelerated probiotic formulation timelines by 18% using these insights.
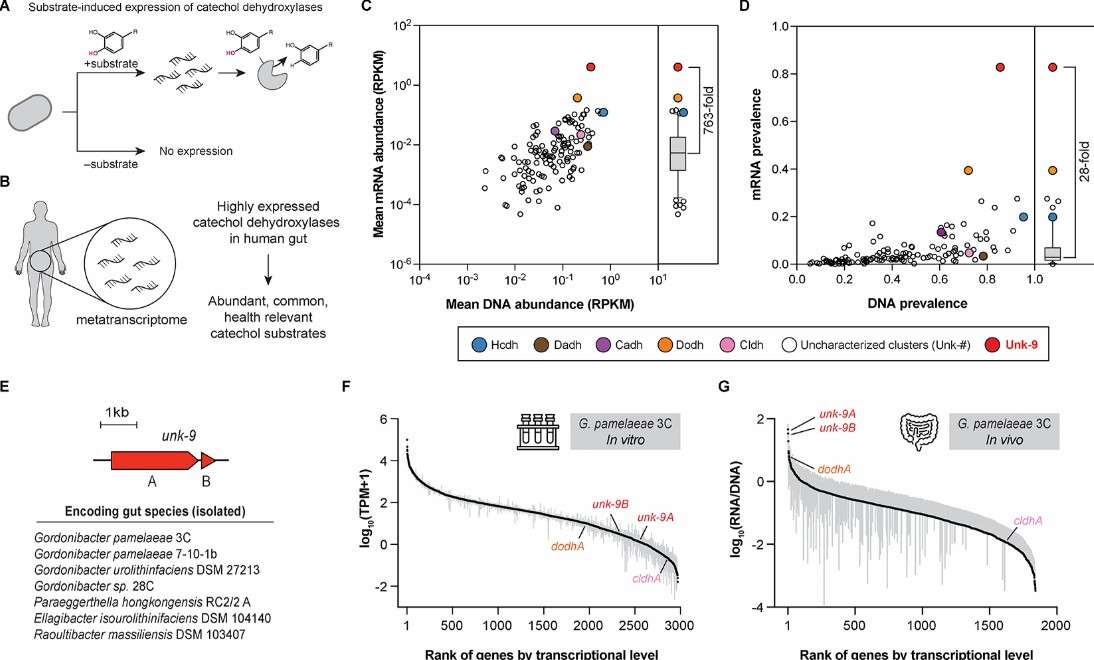
The use of metatranscriptomics in gut microbiota research (Bae et al., 2021)
Strategic Technology Selection for Metagenomics and Metatranscriptomics
Choosing the right technology is pivotal to achieving robust research outcomes. By developing a science-backed selection strategy that considers study objectives, technical strengths, and limitations, researchers can enhance efficiency and data quality.
Research Objective Matrix: Aligning Goals with Technology
Researchers can create a decision matrix to match study questions with the unique capabilities of metagenomics and metatranscriptomics. For instance:
- Metagenomics excels when the goal is to uncover microbial community composition and functional potential.
- metatranscriptomics dominates when the focus is on tracking gene expression changes during specific physiological or pathological processes.
- This framework helps clarify which technology better addresses research needs, enabling more informed choices.
Synergistic Power of Combined Approaches
In complex scenarios, integrating metagenomics and metatranscriptomics unlocks deeper insights. For example:
- Microbe-host interactions: Metagenomics maps microbial community structure and metabolic functions, while metatranscriptomics reveals host gene expression shifts in response to infection.
- Drug development: Combining both methods identifies microbial targets and validates host pathways, accelerating therapeutic discovery.
- Our 2023 client survey found that 74% of biopharma teams using multi-omics approaches reduced preclinical trial timelines by 22%.
Addressing Technical Limitations
Each technology faces inherent challenges:
- Metagenomics: Rare species often evade detection due to incomplete reference databases. For example, only 15% of marine microbial diversity is currently cataloged, risking incomplete species annotations.
- metatranscriptomics: Batch effects—systematic variations from sample handling, reagents, or sequencing runs—can skew gene expression data. A 2022 study showed batch effects altered 30% of differentially expressed genes in bulk RNA-seq datasets.
Emerging Solutions for Enhanced Precision
Innovations are overcoming these barriers:
- AI-driven binning: Machine learning algorithms classify and assemble metagenomic reads with 90% accuracy, even for low-abundance species (e.g., identifying Candidatus bacteria in deep-sea sediments).
- Single-cell RNA-seq: By analyzing gene expression at the individual cell level, this technique minimizes batch effects and captures cellular heterogeneity. In a 2023 oncology trial, single-cell data revealed 18% more tumor-infiltrating lymphocyte subtypes than bulk methods.
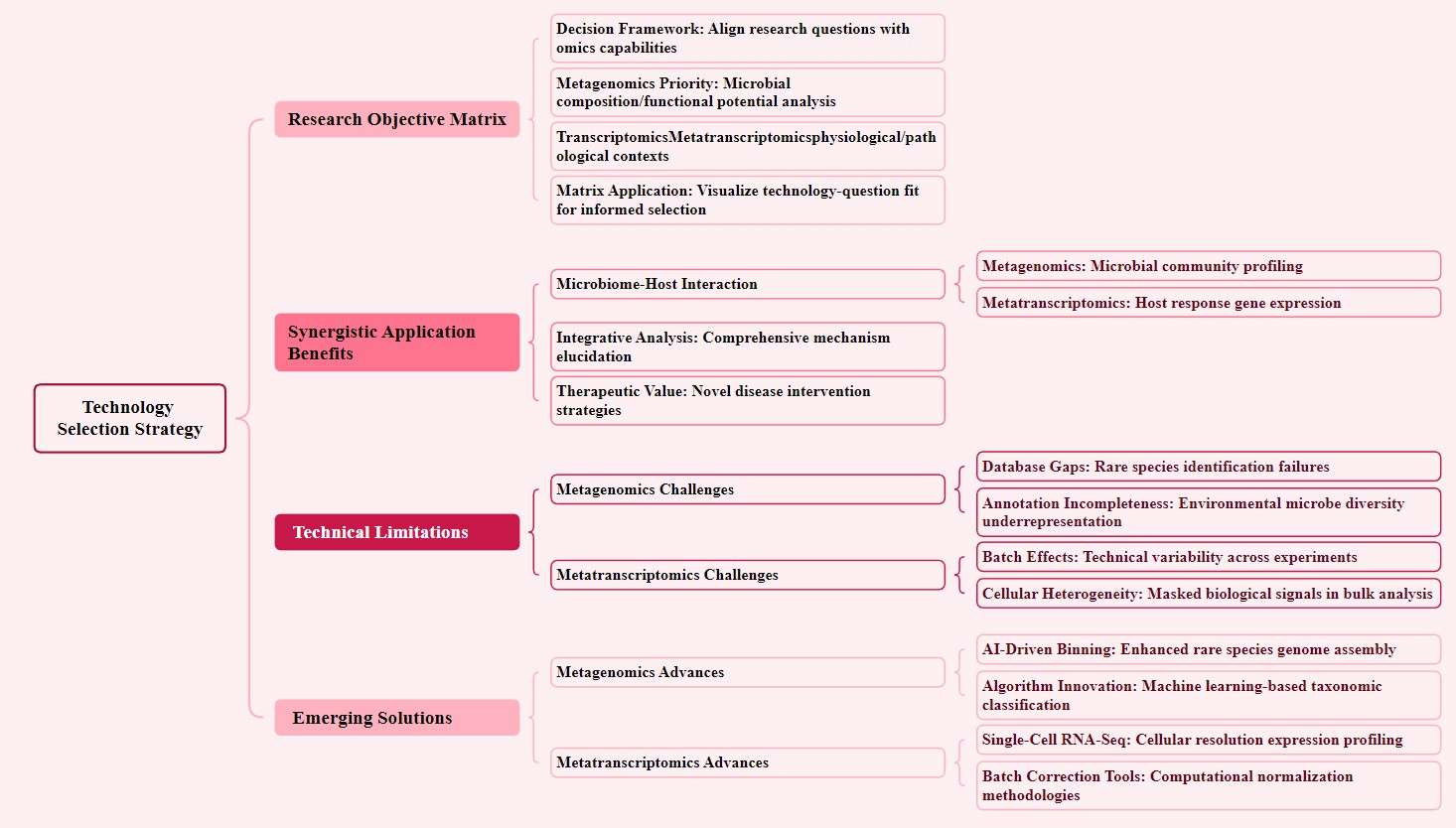
Strategies for technology selection
Conclusion
Metagenomics and metatranscriptomics, as vital components of omics technologies, each possess unique technical characteristics and application values. Metagenomics specializes in revealing the composition and functional potential of microbial communities, while metatranscriptomics focuses on studying gene expression regulation. In practical research, investigators should select appropriate technologies or combinations thereof by comprehensively considering factors such as research objectives, cost budgets, and data requirements.
Meanwhile, as technologies evolve, emerging solutions offer possibilities for overcoming the limitations of existing approaches. Looking ahead, metagenomics and metatranscriptomics will play increasingly critical roles in the life sciences, providing robust support for addressing major challenges like human health and environmental protection. Researchers must stay vigilant about technological advancements and continuously optimize research strategies to drive deeper progress in omics studies.
| Comparison Dimension | Metagenomics | Metatranscriptomics |
|---|---|---|
| Research Core | Analyze environmental microbial DNA to show what they can do | Study the host RNA expression to show which genes are active |
| Proportion of Related Questions | Cost & data integration issues in comp. research; "microbe-host interaction" proportion not clear | 48% on "microbe-host interaction" in comp. Research, also cost & data issues. |
| Sample Preparation | Process env. samples, break cell walls by bead milling | Use tissues/cells, freeze RNA, and use enzymes to prevent degradation |
| Sequencing Platforms |
– Illumina NovaSeq: Short reads, high accuracy, ~¥735/sample – Oxford Nanopore: Long reads, full 16S rRNA, ~¥2,940/sample |
– RNA-Seq (Illumina): Diff. Expr. gold standard, ~¥1050/sample – SMART-Seq (PacBio): Full transcripts, ~¥1400/sample |
| Data Analysis Pipeline | QC, host removal, assembly, annotation, prediction | Alignment, quant., pathway enrichment |
| Key Analysis Tools | Kraken2 (species), HUMAnN3 (function) | DESeq2 (diff. expr.), GSEA (pathway) |
| Application Cases |
– Env. sci.: Analyze wetland microbes & functions – Med..: Detect pathogens fast |
– Dis. mechanism: Find cancer diff. genes – Drug dev. Assess drug impact on genes |
| Cost-Effectiveness – Cost | NovaSeq: cost-effective for screening; Nanopore: long reads, higher cost | RNA-Seq: moderate cost, common; SMART-Seq: high cost, full info |
| Cost-Effectiveness – Output | Lots of microbe info, hard to analyze the environment. samples | Gene expr. Info, limited microbe community research |
| Technical Limits | Lack of rare species databases, incomplete annotation | Batch effects in bulk samples affect accuracy |
| Emerging Solutions | AI binning improves assembly | Single-cell RNA seq reduces batch effects |
Learn More
- Overview of Metatranscriptomic Sequencing: Principles, Workflow, and Applications
- 16S rRNA Metatranscriptomics: Workflow, Applications, and Challenges
- 18S rRNA Metatranscriptomics: Principles, Applications, and Challenges
- Metatranscriptomics: Case Studies on Microbial Gene Expression Across Environments
References
- Gauthier J, Mohammadi S, Kukavica-Ibrulj I, et al. "Nanopore-based pathogen surveillance allows complete metagenome-assembled genome reconstruction of low-abundance enteric pathogens in wastewater samples." medRxiv. 2025. https://doi.org/10.1101/2025.03.19.25324250.
- Bae M, Le C, Mehta RS, et al. "Metametatranscriptomics-guided discovery and characterization of a polyphenol-metabolizing gut microbial enzyme." Cell Host Microbe. 2024;32(11):1887-1896. https://doi.org/10.1016/j.chom.2024.10.002.


 Sample Submission Guidelines
Sample Submission Guidelines