Scientists can now study the complete set of DNA—the genome—from just one cell. This is a big step in biology and could change how we find and treat diseases. A key part of this is Single-Cell Whole-Genome Amplification (WGA). Think of WGA as a way to make many copies of the tiny amount of DNA in a single cell so it can be studied. This article will explain why WGA is important, what makes studying single cells tricky, how WGA methods have gotten better, how it’s used in research, and what challenges and future improvements we might see.
Why WGA Is Essential in Single-Cell Genomics
Our genome is like a blueprint for life. It’s different and changes, especially when we look at it one cell at a time. WGA helps us read this detailed blueprint.
The Value of Single-Cell Genomes
Finding Cell Differences: In our bodies or in groups of tiny living things like bacteria, each cell can have its own unique DNA features. WGA helps scientists study these differences. This allows them to track how cells are related, how they grow and develop, and how groups of cells change over time, like in growing tumors or bacterial colonies.
Studying Rare and Special Cells: Sometimes scientists need to study cells that are hard to find, like cancer cells floating in the blood (CTCs), egg cells, or certain stem cells. Other times, each cell’s DNA is naturally unique, like in sperm cells after DNA shuffling (meiosis) or as cells age and gather DNA changes. WGA makes enough DNA from these single, special cells so scientists can study them fully, looking at things like cancer differences or the genetic health of reproductive cells.
A New View of DNA Changes: Looking at DNA from single cells with WGA gives us a new way to see how DNA varies between cells. This is very helpful in cancer research to map how tumors grow and change. It’s also useful in understanding how our bodies develop, by tracking DNA changes as cells specialize to do different jobs.
Amplifying Small Amounts of DNA
Tiny Amounts of DNA: A single cell from a mammal has only a tiny bit of DNA, about 6 to 10 trillionths of a gram. This isn’t nearly enough for most DNA testing machines, which need much more. So, making many copies of the entire genome is a must before any testing can be done, like with next-generation sequencing (NGS).
WGA Makes Study Possible: WGA is the vital step that makes single-cell DNA study possible. It closes the gap between the tiny amount of DNA in one cell and what modern tools need. It’s the foundation of single-cell DNA research, overcoming the small amount of DNA to unlock a lot of genetic information.
Challenges and Needs in Single-Cell WGA
Working with single cells is tough. From catching one cell to checking the quality of copied DNA, WGA methods have to be very good.
The Difficulty of Isolating Rare Cells: Getting just one target cell, especially if it’s rare (less than 1 in 100 cells in a mix), is a big challenge. If DNA from other cells or the environment gets in, it can mess up the results. This means scientists need very careful ways to isolate cells.
Critical Performance Imperatives for WGA
The success of any single-cell DNA study depends on how well the WGA method works. Key things to look for are:
- Genome Coverage: How much of the cell’s total DNA gets copied and read. High coverage is needed to find all the DNA changes.
- Amplification Uniformity: Ideally, all parts of the DNA should be copied equally. If not, some parts get over-copied and others under-copied or missed, which creates errors.
- Amplification Fidelity: The copying process shouldn’t make many mistakes (like wrong DNA letters). High accuracy is key for finding small DNA changes called SNVs.
- Allele Dropout (ADO) Rate: We get two versions (alleles) of most genes, one from each parent. ADO happens if one version isn’t copied. Low ADO is important for correct genetic typing.
- Chimera and Unmappable Read Rates: Chimeras are jumbled DNA pieces made by mistake during copying. Too many of these, or DNA pieces that can’t be matched to the original genome, make the data hard to use.
- Reproducibility: The WGA method should give the same results if repeated on similar cells or in different lab runs.
- Contamination Control: Since there’s so little starting DNA, it’s easy for unwanted DNA (from the lab or reagents) to get in. Strict steps are needed to prevent this.
Improving WGA: Better Ways to Copy Single-Cell DNA
Scientists have worked hard to make Single-Cell Whole-Genome Amplification better and more reliable.
Older WGA Methods: How They Work and Their Limits
- DOP-PCR (Degenerate Oligonucleotide-Primed PCR):
How it Works: Uses primers (short DNA pieces that start the copying) with a mix of random sequences at one end and a set sequence at the other. Copying starts at low temperatures, then uses higher temperatures.
Pros & Cons: Useful for seeing large DNA changes (Copy Number Variations, or CNVs). But, it often doesn’t copy the whole genome and can copy unevenly because PCR makes copies exponentially (like a snowball effect).
- MDA (Multiple Displacement Amplification):
How it Works: Uses an enzyme called phi29 DNA polymerase, which is good at making long DNA copies and can push existing DNA strands out of the way. It works with random short primers at a steady temperature.
Pros & Cons: Usually copies more of the genome and makes fewer mistakes than PCR methods. But, it can still copy unevenly, especially with the tiny bit of DNA from one cell. The basic phi29 enzyme isn’t always efficient enough for single cells.
- MALBAC (Multiple Annealing and Looping-Based Amplification Cycles):
How it Works: Designed to reduce uneven copying. It does a few cycles of almost linear (one-by-one) copying first, using primers that make the copied DNA form loops. These loops stop those pieces from being re-copied too early, making it more even. Then, regular PCR is used.
Pros & Cons: Better at copying evenly and finding small differences than MDA. Good for CNV analysis because its unevenness is more predictable.

Figure 1. Traditional single-cell WGA methods.(Huang L. et al, 2015)
Newer WGA Methods: Big Improvements
Scientists developed new WGA techniques for better results:
- PTA (Primary Template-Directed Amplification):
How it Works: Uses the accurate phi29 enzyme but adds special DNA building blocks that stop the copying process after a certain length. This helps control the size of the copied DNA pieces made directly from the cell’s original DNA, leading to more even copying across the whole genome.
Benefits: PTA copies more of the genome more evenly, almost as well as methods using much more DNA. It’s very good at finding small DNA changes (SNVs, reportedly over 90% found) and greatly reduces missed alleles (ADO) and uneven copying.
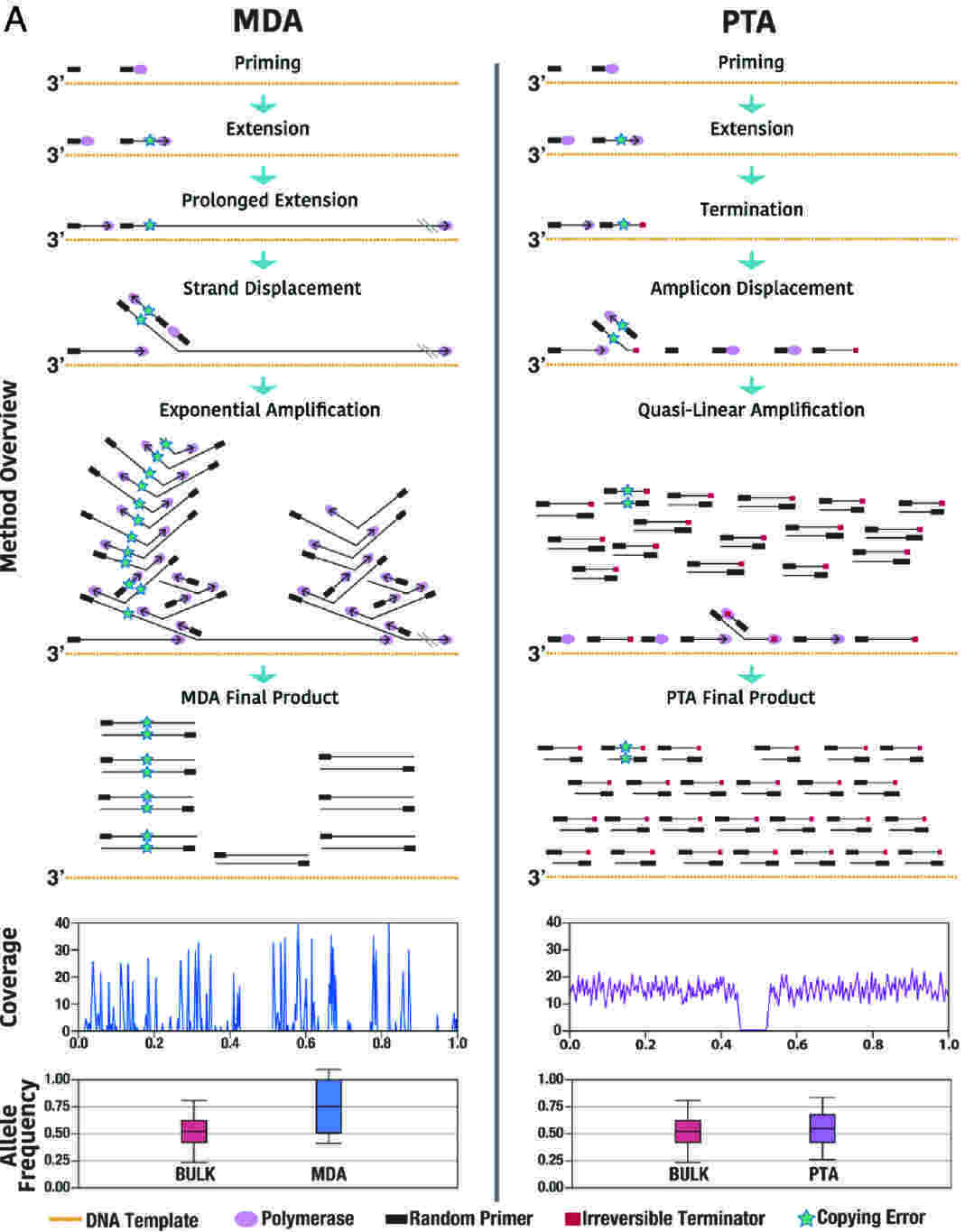
Figure 2. Single-cell WGA methods between MDA and PTA.(Gonzalez-Pena V. et al, 2021)
- iSGA (Improved Single-cell Genome Amplification):
How it Works: This method improves MDA by changing both the phi29 enzyme and the copying process.
Enzyme Changes: Scientists made the phi29 enzyme more stable and active at higher temperatures (around 40°C) by adding things like disulfide bonds (making "HotJa Phi29"). They also made other changes and added helper proteins to get more of the enzyme and make it work better.
Process Changes: They use UV light to clean reagents of any stray DNA, adjusted the chemical mix (like potassium levels), and added substances to keep the enzyme stable.
Benefits: iSGA with the changed enzyme works very well at 40°C, copies a very high percentage of the genome (up to 99.75% in some tests), is reliable, and costs less than many commercial kits.
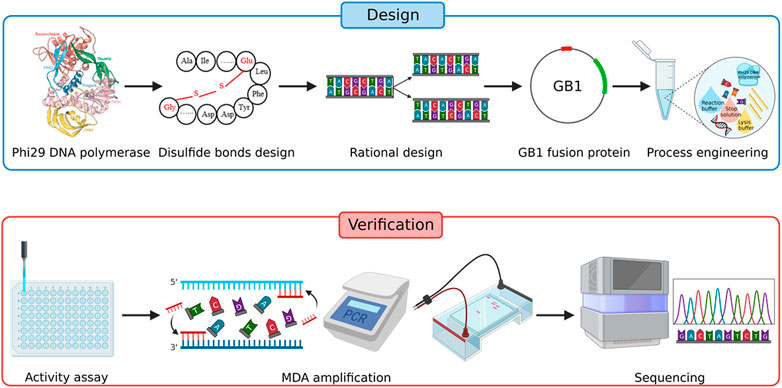
Figure 3. Strategy for improving the amplification efficiency of phi29 DNA polymerase.(Zhang J. et al, 2023)
Applications of WGA in Pre-Clinical Research and Scientific Discovery
Copying DNA from single cells has opened up many research areas. Remember, these are research uses, not for diagnosing medical conditions.
Cancer Studies
- Understanding Tumor Growth: WGA helps scientists see how cancer cells change over time, find key DNA changes in different cells, and understand how complex tumors are. This information is vital for creating better, more targeted cancer treatments.
- Studying Circulating Tumor Cells (CTCs): CTCs are rare cancer cells that break off from tumors and travel in the blood. WGA allows scientists to study the DNA of these few cells, giving clues for predicting outcomes and guiding treatment research.
Reproduction and Development Research
- Preimplantation Genetic Diagnosis (PGD) and Screening (PGS) Research: In IVF research, WGA is used on single cells from embryos to check for chromosome problems (PGS) or specific genetic mutations (PGD). This helps understand what makes embryos healthy and aims to prevent genetic diseases.
- Studying Egg and Sperm Cells: WGA helps study DNA recombination (shuffling) and DNA health in individual human eggs and sperm. This gives a better understanding of fertility, infertility, and how new genetic conditions can arise.
- How Embryos Develop: By looking at the DNA of single cells at different growth stages, scientists can learn about the genetic plans that guide how embryos form and cells specialize.
Immune System and Microbe Research
- Understanding Immunity: WGA can be used on single immune cells (like lymphocytes) to study how our bodies create diverse immune responses and how DNA changes in immune cells might relate to diseases.
- Studying Single Microbes: Most microbes can’t be grown in labs. WGA is key for studying the DNA of these single, unculturable bacteria, understanding microbe diversity, how germs evolve, and what individual microbes do in complex groups (like in our gut).
Current Hurdles and What’s Next for Single-Cell WGA
Even with big improvements, Single-Cell Whole-Genome Amplification still has challenges. Solving these is key to getting the most out of single-cell DNA studies.
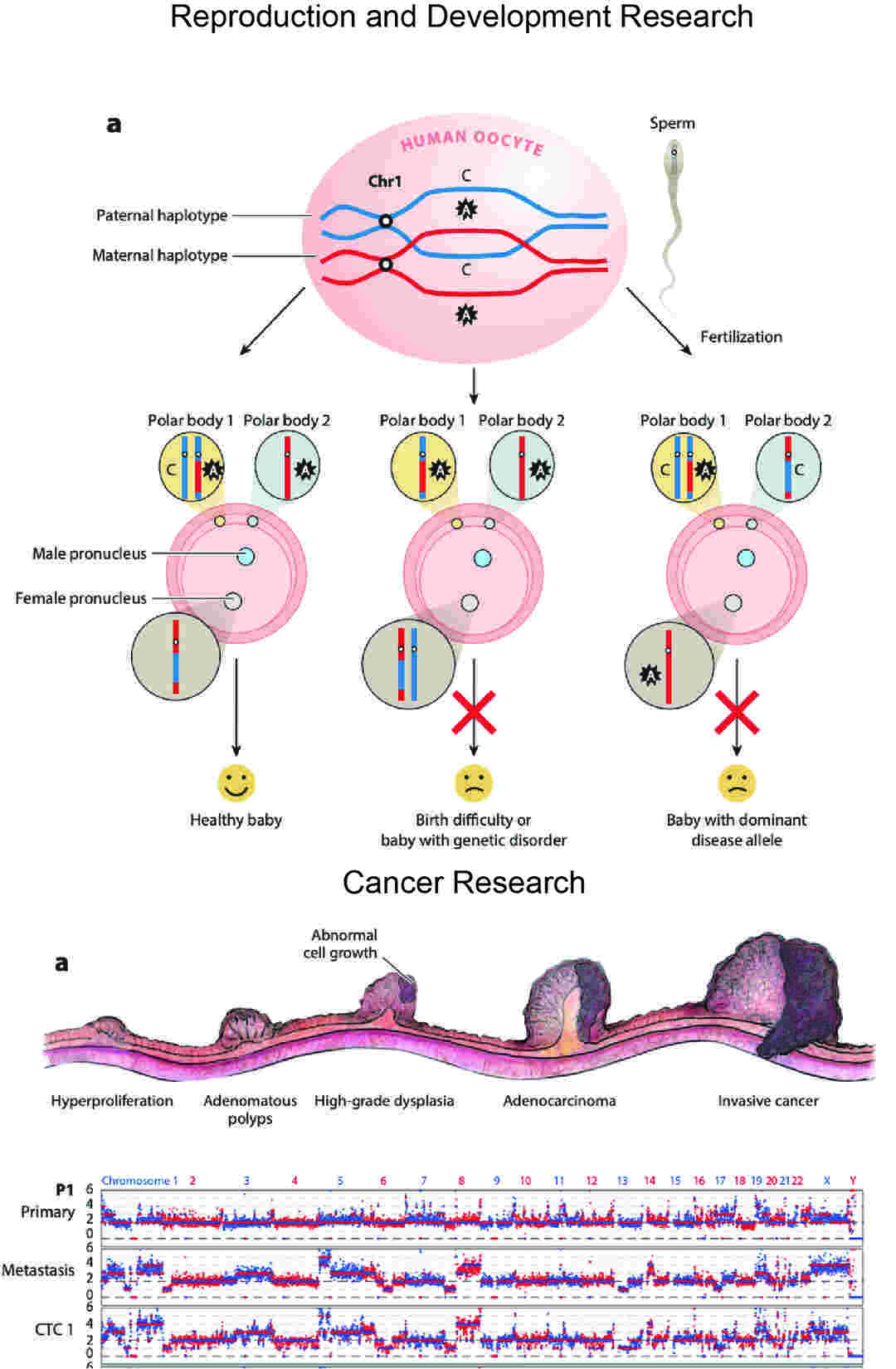
Figure 4. Traditional single-cell WGA methods.( Huang L. et al, 2015)
Main Challenges Today
- Uneven and Incomplete Copying: It’s still hard to copy every part of the genome completely and evenly. Some DNA regions (like those with lots of G and C letters, or repetitive parts) get copied too much or too little.
- Copying Errors and Biases: All WGA methods make some mistakes, like putting in the wrong DNA letter, creating jumbled DNA pieces, or copying one parent’s gene version more than the other (which can lead to allele dropout).
- Copying Enough from Tiny Amounts: Getting enough copied DNA from the tiny bit in one cell is tough. If not enough is copied, the genome picture is incomplete.
- Contamination: Because there’s so little starting DNA, tiny amounts of stray DNA from the lab or reagents can easily contaminate the sample and give wrong results.
- Complex Data Analysis: The copied DNA data has unique errors and biases. Special computer programs are needed to correctly find DNA changes, CNVs, and understand the results. Regular DNA analysis tools might not be good enough.
Future Trends
The field is moving fast to improve WGA:
- Better Enzymes: Scientists are still working to make DNA copying enzymes (like phi29) even better – more accurate, faster, able to copy more DNA, and less biased.
- New WGA Methods: The search is on for WGA methods that copy everything evenly, make almost no mistakes, and are easy to use, perhaps using new chemistry or ideas beyond current PCR and MDA.
- Faster, Cheaper Processes: Making WGA workflows simpler, using robots, and doing reactions in even smaller volumes (like with microfluidics) will allow more cells to be studied, lower costs, and give more consistent results. This will help with big projects like mapping all human cell types.
- Combining with Other ‘Omics’: The real power is in getting many types of information from the same single cell. Future methods will combine WGA with ways to study RNA (gene activity), epigenetics (DNA modifications), and proteins. This will give a complete picture of what cells are doing.
- Smarter Computer Tools: Better computer programs are vital. Future software will be better at fixing errors, removing biases, accurately finding all types of DNA changes, and making sense of complex single-cell DNA data.
- More Research Uses: As WGA gets better and easier, it will be used in even more areas of basic research. Making methods standard and well-tested will also be key for any future (non-diagnostic) uses in more regulated settings.
Conclusion
Single-Cell Whole-Genome Amplification is a vital technology in modern biology. It lets scientists study the DNA details of individual cells like never before. The progress from early methods like DOP-PCR to advanced ones like PTA and iSGA shows a history of smart innovation to make DNA copying more accurate, even, and complete. Big challenges related to bias, errors, and efficiency remain, but ongoing work in enzyme improvement, new methods, and data analysis promises an even more powerful future for WGA.
The many uses of WGA—from understanding cancer and reproduction to checking environmental effects and gene editing—show its huge impact on our knowledge of life at the cellular level. As these technologies keep improving, they will surely give us more insights into how cells work, how diseases develop, and how life evolves. This makes the single cell a key focus for future discoveries. The ongoing effort to create the "perfect" WGA method is essential for the next big breakthroughs in genomics.
References
- Huang L, Ma F, Chapman A. et al. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu Rev Genomics Hum Genet. 2015;16:79-102. https://doi.org/10.1146/annurev-genom-090413-025352
- Gonzalez-Pena V, Natarajan S, Xia Y. et al. Accurate genomic variant detection in single cells with primary template-directed amplification. Proc Natl Acad Sci U S A. 2021 Jun 15;118(24):e2024176118. https://doi.org/10.1073/pnas.2024176118
- Zhang J, Su X, Wang Y. et al. Improved single-cell genome amplification by a high-efficiency phi29 DNA polymerase. Front Bioeng Biotechnol. 2023 Jun 29;11:1233856. doi: 10.3389/fbioe.2023.1233856. Erratum in: Front Bioeng Biotechnol. 2023 Aug 28;11:1263634. https://doi.org/0.3389/fbioe.2023.1263634


 Sample Submission Guidelines
Sample Submission Guidelines