Fungi are a kind of magical creatures. They are in various forms, from microscopic yeasts to mushrooms and fungus visible to the naked eye. It is not only different from the photosynthesis of plants, but also different from the feeding mode of animals. It survives by absorbing nutrients from the surrounding environment. Widely distributed in nature, it plays a key role in the material cycle of the ecosystem, and some of them are closely related to human life, or used in food or medicine.
This paper introduces the basic characteristics of fungi and their relationship with human beings, summarizes the principle, application and future prospect of fungal sequencing technology.
Overview of Fungi
Fungi are a kind of eukaryotes, which exist widely in nature. They are diverse, ranging from single-celled yeasts to multicellular molds and large fungi such as mushrooms. Fungi have no chlorophyll, can’t carry out photosynthesis, and get nutrition by absorbing organic substances in the surrounding environment. Its reproduction methods include asexual reproduction and sexual reproduction. In the ecosystem, fungi play an important role, which can decompose organic matter and promote material circulation. Some fungi can be used in food fermentation and pharmacy, but some can also cause animal and plant diseases.
Basic Feature
Cell structure: Fungi are eukaryotic microorganisms with highly differentiated nuclei, including nuclear membranes and nucleoli. There are complete organelles in the cytoplasm, and the cell wall is made of chitin or cellulose, which has no chlorophyll and does not differentiate roots, stems and leaves.
Morphology and type: A few are single cells, and most are multicellular. Single-celled fungi, such as yeast-like and yeast-like fungi, are round or oval; Multicellular fungi are composed of hyphae and spores, such as filamentous fungi.
Survival and Reproduction
Life style: It is widely distributed in nature and has many kinds. It lives in saprophytic or parasitic way, and can live in soil, water, animals and plants, their remains and air.
Reproductive methods: Including asexual reproduction and sexual reproduction, and quasi-sexual reproduction also exists. Asexual reproduction includes somatic cell rupture, division, budding and the production of asexual spores. Sexual reproduction takes the form of gametophyte mating and gametophyte mating, and finally forms sexual spores such as dormant sporangium and oospore.
Main Classification
Single-celled fungi: Such as yeast fungi, which do not produce hyphae, germinate and reproduce through mother cells, and the colonies are similar to bacterial colonies; The colony of yeast-like fungi is similar to that of yeast, but there are pseudohyphae formed in the culture medium.
Multicellular fungi: They are composed of hyphae and spores, such as filamentous fungi and Candida albicans. The hyphae can form different structures, such as funiculus, sclerotia and stroma, while spores are reproductive structures.
Relationship with Human Beings
Beneficial aspects: Many fungi can be used for brewing and fermentation, and also can produce antibiotics and other drugs, such as penicillin, which is produced by Penicillium. In addition, some fungi are delicious food materials, such as mushrooms and auricularia, and some fungi play an important role in decomposing organic matter and promoting material circulation in the ecosystem.
Harmful aspects: Some fungi can invade the human body and cause infection, causing skin itching, blisters, fever, cough and other discomfort. In severe cases, it can cause sepsis, septic shock, multiple organ failure and even death, such as pulmonary candidiasis and cryptococcal meningitis.
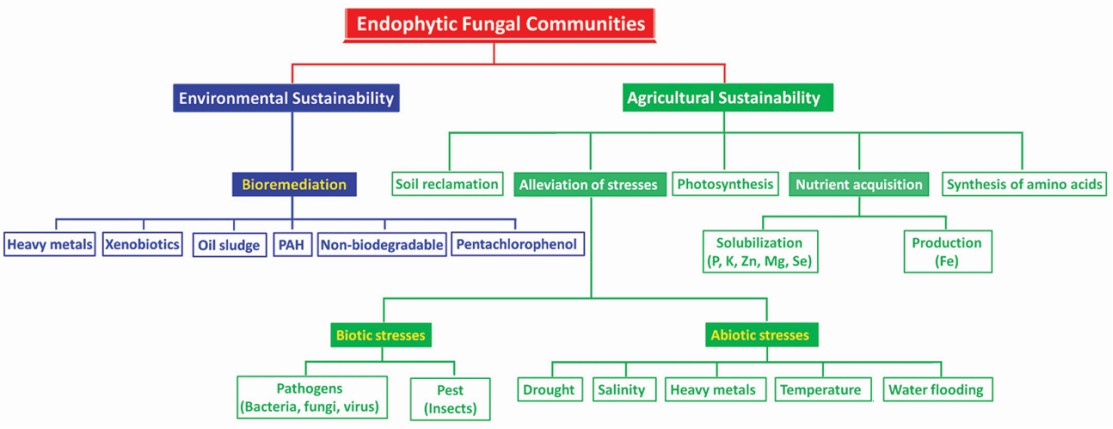
Biotechnological applications of endophytic fungal communities for agro-environmental sustainability (Kour et al., 2023)
Basis of Fungal Sequencing Technology
Fungal sequencing technology is an important means to study fungi, which mainly includes obtaining fungal genome sequence based on the second-generation high-throughput sequencing technology, and performing genome component analysis, functional annotation and other analysis. It can be used to predict the function of fungal genes and proteins, identify the interaction between pathogenic fungi and crops and pathogenic genes, study evolutionary relationships and compare genomes, and discover fungal metabolic pathways and beneficial metabolites.
Fungal Genome Sequencing
Fungal genome sequencing refers to sequencing the whole genome of fungi to obtain its complete DNA sequence information. By comparing the genome sequences of different fungi, it can reveal their genetic relationship and evolutionary process, and provide an important basis for the classification and phylogenetic study of fungi. For plant and human pathogenic fungi, genome sequencing is helpful to discover genes and metabolic pathways related to pathogenicity, deeply understand the pathogenic mechanism of fungi, and provide theoretical basis for disease prevention and treatment.
Understanding the genomic information of fungi can help to find new drug targets and develop new antifungal drugs for fungi to cope with the increasingly serious fungal infection. For some fungi used in industrial production, such as Saccharomyces cerevisiae and enzyme-producing fungi, genome sequencing can provide a target for genetic modification, and improve their fermentation performance, product yield and quality through genetic engineering technology.

DNA barcoding (A) vs DNA taxonomy (B) (adapted from Vogler and Monaghan) (Raja et al., 2017)
Fungal Transcriptome Sequencing
The sequencing of fungal transcriptome is to high-throughput sequencing all RNA transcribed by fungi under a certain condition, so as to obtain the information of fungal transcription level comprehensively and quickly. Through transcriptome sequencing, it can find new genes in fungi and annotate their functions, which is helpful to improve the genomic information of fungi. To study the changes of gene expression of fungi in different growth stages or development processes and understand the molecular mechanism of their growth and development. It can also analyze the differences of gene expression of fungi under biological stress (such as pathogen infection) or abiotic stress (such as temperature, humidity, pH change, etc.), reveal its molecular regulatory network to deal with stress, and provides theoretical basis for improving the stress resistance of fungi.
Fungal transcriptome sequencing has significant advantages. The gene expression profile can be comprehensively analyzed and functional genes can be mined. It can be carried out without reference genome and discover new transcripts. It helps to study the growth and development, pathogenic mechanism, response to stress and other processes of fungi, with high flux and accurate data, providing key information and theoretical basis for fungal genetic improvement, drug research and development and disease prevention and control.
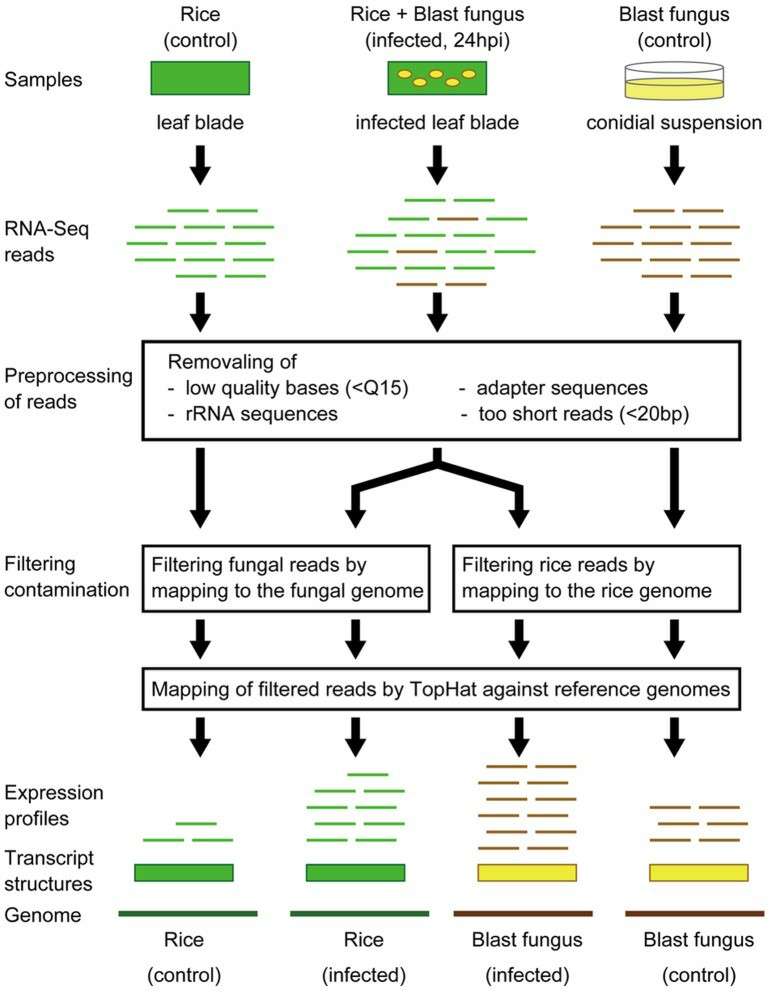
Schematic representation of RNA-seq analysis of mixed transcriptome obtained from blast fungus-infected rice leaves (Kawahara et al., 2012)
Fungal Metagenome Sequencing
Fungal metagenome sequencing is to directly extract the DNA of all fungi from environmental samples (such as soil, water, biological tissues, etc.) without relying on traditional pure culture technology, and then construct a metagenome library, and then sequence the DNA fragments in the library by using high-throughput sequencing technology, so as to obtain the genomic information of fungal communities in the environment.
Metagenome sequencing is mainly used to study the structure and diversity of fungal communities in soil, ocean, fresh water and other environments, as well as their functions in ecosystems, such as material circulation and energy conversion. It is helpful to study the composition and changes of fungi in human microbiome and their relationship with diseases. Through metagenome sequencing, we can deeply understand the pathogenesis of these diseases and provide new ideas for diagnosis and treatment. It can be used to study the relationship between plant diseases and fungal communities, and the promotion of beneficial fungi in soil to plant growth.
Specific Fungal Sequencing Technology
18S rRNA gene sequencing: 18S rRNA gene exists in eukaryotic ribosomes, which is highly conserved and changes slowly during evolution. It can provide a strong basis for the classification and identification of fungi, and can be used to determine the attribution of fungi in higher classification order such as boundary, phylum, class and order. By sequencing the 18S rRNA gene, we can understand the genetic relationship between different fungal groups, construct a reliable phylogenetic tree, and help researchers explore the evolutionary process of fungi. When studying unknown fungi, the 18S rRNA gene sequence can be compared with the known fungi sequence, and the category can be preliminarily judged. However, this gene is highly conservative, and its resolution is insufficient in distinguishing closely related fungal species.
Internal Transcribed Spacer (ITS) sequencing: ITS is located in the transcription unit of fungal ribosomal RNA gene and is divided into ITS1 and ITS2 regions. Compared with the 18S rRNA gene, the sequence variability of ITS region is higher, which can reflect the differences within and among true strains. This feature makes ITS sequencing the "gold standard" for fungal species identification, and it is widely used in fungal classification and identification. ITS sequencing can accurately distinguish different species when identifying some fungi which are similar in morphology and difficult to distinguish. In addition, when studying the structure and diversity of fungal communities, ITS sequencing can detect the species and relative abundance of fungi in environmental samples, and understand the composition and distribution of fungal communities. However, it also has limitations. In some complex fungal groups, sequence polymorphism may lead to difficulties in analysis.
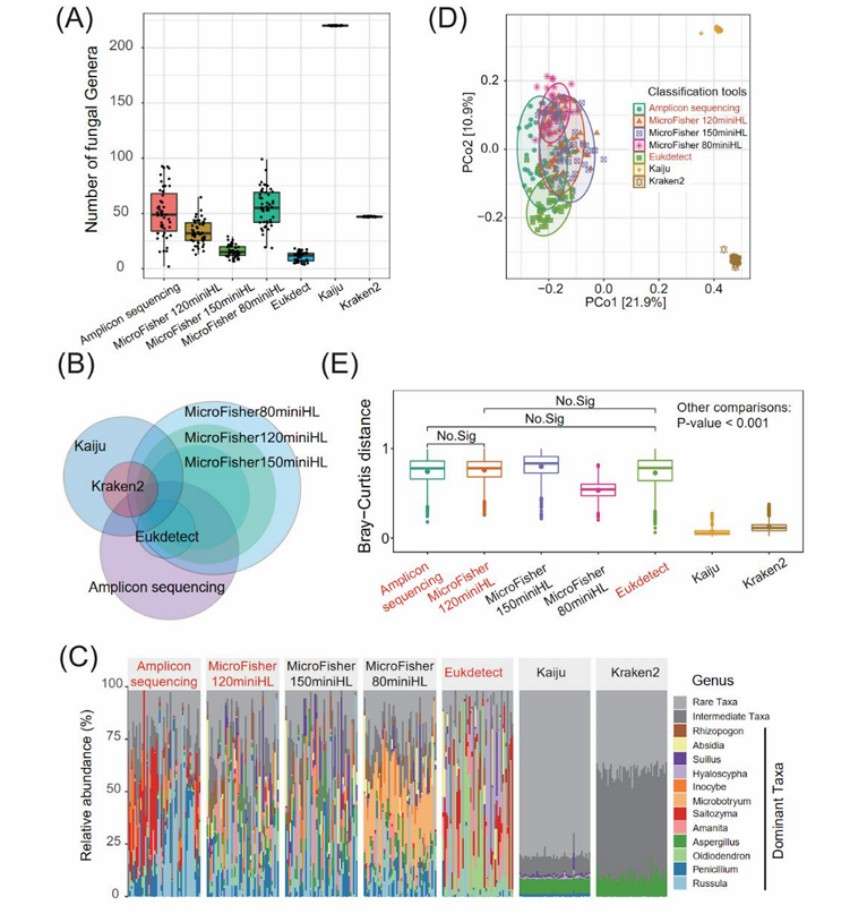
The analysis of soil metagenome data using MicroFisher versus other fungal classification tools (Wang et al., 2024)
Fungal Sequencing Application
In fungal research, sequencing technology has a wide range of applications, mainly including the following aspects.
Study on Taxonomy and Phylogeny of Fungi
Accurate identification of species: Traditional fungi classification is mainly based on morphological characteristics, but many fungi are similar in morphology, so it is difficult to distinguish them accurately. By analyzing the ribosomal DNA (rDNA) sequence of fungi, such as the internal internal transcribed spacer (ITS) sequence, sequencing technology can accurately identify species or even subspecies. For example, in some Aspergillus fungi with similar morphology, different species can be clearly distinguished by ITS sequence analysis.
Construction of phylogenetic tree: Based on multiple genes or whole genome sequences, the evolutionary relationship between fungi can be understood more comprehensively and an accurate phylogenetic tree can be constructed. For example, by sequencing the genomes of many basidiomycetes, their phylogenetic relationships and evolutionary branches are revealed, which provides a more reliable basis for the systematic classification of fungi.
Study on the Structure and Function of Fungal Genome
Genome sequencing and assembly: With the development of sequencing technology, more and more fungal genomes have been completely sequenced and assembled. For example, the genome of Saccharomyces cerevisiae is the first eukaryotic genome to be completely sequenced, which provides an important reference for the follow-up fungal genome research. Through genome sequencing, we can understand the basic information of fungi, such as gene composition and chromosome structure, and lay a foundation for further study of their biological functions.
Gene annotation and function prediction:The genome sequence obtained by sequencing is annotated by bioinformatics method to predict the function of the gene. For example, by comparing with gene sequences with known functions, we can infer the role of some unknown genes in the growth, development and metabolism of fungi. At the same time, combined with the technology of RNA-seq, the expression of the gene under different growth conditions was analyzed to further verify the function of the gene.
Genetic Breeding of Fungi
Mining excellent gene resources: Sequencing fungi with excellent characters to find genes related to high yield, high quality and stress resistance. For example, in some fungi producing antibiotics, gene clusters related to antibiotic synthesis were found by genome sequencing, which provided gene resources for further improving antibiotic production and developing new antibiotics.
Assisted breeding technology: Using sequencing technology to carry out molecular marker-assisted selection breeding can quickly and accurately screen out strains with target traits. For example, in the breeding of Lentinus edodes, molecular markers related to the quality and yield of Lentinus edodes were developed by sequencing the genomes of different strains, and these markers were used for screening in the breeding process, which greatly improved the breeding efficiency.
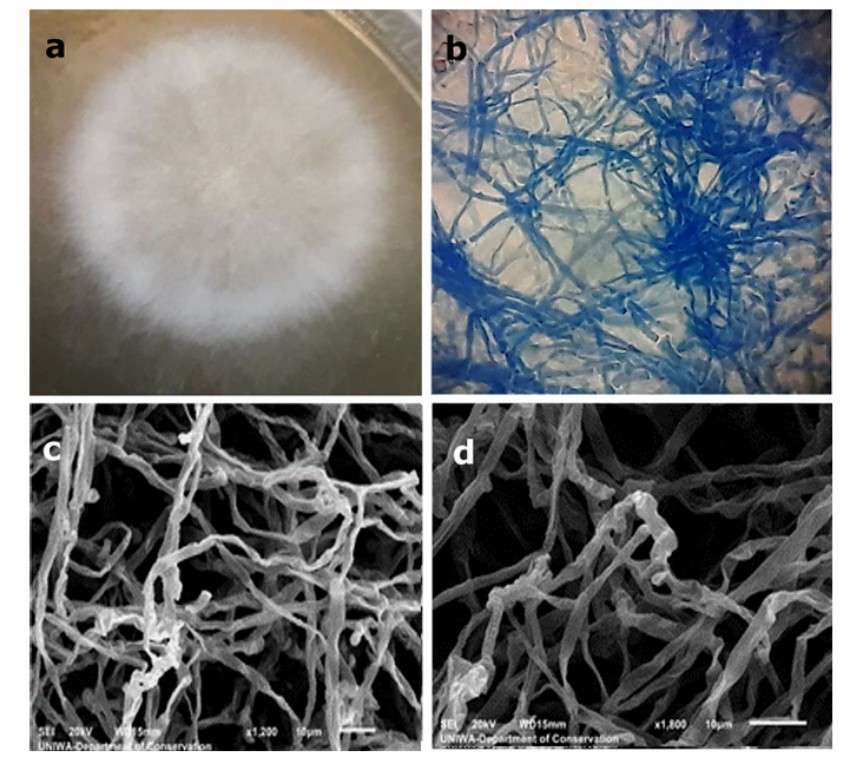
Observation of the mycelium of the unknown fungus (Kokla et al., 2023)
The Future Prospects of Fungal Sequencing
With the rapid development of biotechnology, fungal sequencing, as a powerful tool to explore the mysteries of the fungal world, is at a critical stage of continuous innovation and application, and its future prospects are full of infinite possibilities.
In the field of basic research, fungal sequencing will help to fully reveal the molecular mechanism of fungal life activities. On the one hand, by sequencing the whole genomes of a large number of fungi with different niches and functions, a huge and fine fungal genome database is constructed, which is helpful to deeply understand the evolutionary process of fungi, clarify the genetic relationship between species and adapt to environmental changes. On the other hand, combining with the technologies of transcriptomics, protein omics and metabonomics, the expression regulatory network of fungal genes was analyzed in different temporal and spatial dimensions, and the molecular events in the process of fungal growth, development, reproduction and interaction with other organisms were comprehensively explained.
In clinical diagnosis, fungal sequencing has a broad application prospect. Next-generation sequencing of metagenome (mNGS) is expected to become a routine method for clinical diagnosis of fungal infection. By unbiased detection of all nucleic acids in patients’ samples, pathogenic fungi, including rare and new pathogenic fungi, can be quickly and accurately identified, which greatly shortens the diagnosis cycle and strives for valuable treatment time for patients. At the same time, with the optimization of bioinformatics algorithm and the continuous improvement of database, the interpretation of mNGS results will be more accurate and reliable, effectively reducing false positive and false negative results and improving the accuracy of clinical diagnosis.
In the agricultural field, fungal sequencing will inject new vitality into crop disease prevention and the development of edible fungi industry. For plant pathogenic fungi, sequencing technology can be used to identify their pathogenic genes, clarify the interaction mechanism between pathogens and crops, and then develop accurate prevention and control strategies based on gene targets. In the study of edible fungi, by sequencing the genome of edible fungi, genes related to the synthesis of nutrients, the metabolism of flavor substances and stress resistance can be mined, which provides theoretical basis for the variety improvement and high-quality and efficient cultivation of edible fungi and promotes the development of edible fungi industry in the direction of high quality and high yield.
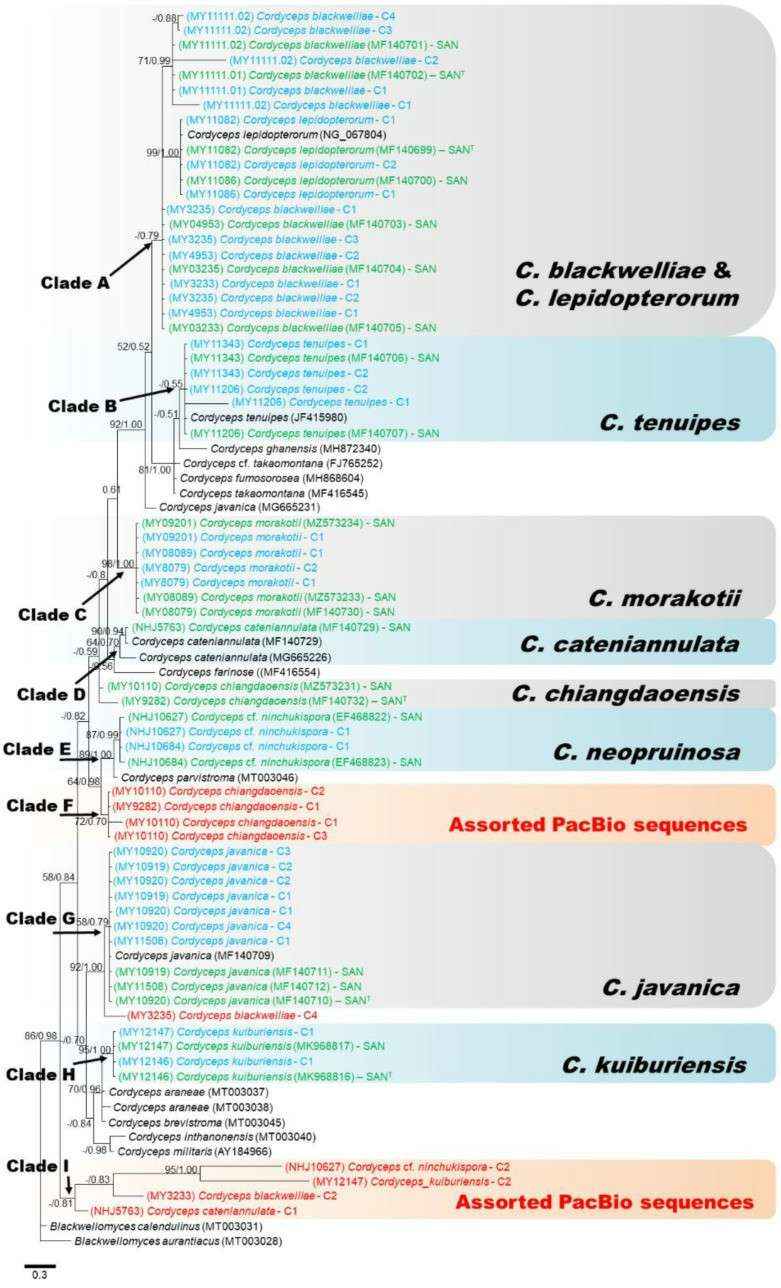
Consensus phylogram (50% majority rule) from a Bayesian analysis of the nrLSU sequences, obtained from 106 MCMC generations (Paloi et al., 2021)
Conclusion
Fungi are a kind of eukaryotes, including yeasts, molds and mushrooms. They play a key role as decomposers in the ecosystem and are closely related to human life. For example, they are used to ferment food, produce antibiotics, and sometimes cause diseases. Fungal sequencing means sequencing and analyzing the genome of fungi. With the development of sequencing technology, the whole genome sequence of fungi can be quickly determined, which helps people to deeply understand the genetic information, evolutionary relationship, pathogenic mechanism and metabolic pathway of fungi. It is of great significance for fungal classification and identification, resource development and utilization, disease prevention and ecological research, and promotes the development of mycology research.
Learn More:
- Introduction to Eubacteria: Taxonomy, Reproduction, Metabolism and Research Methods
- Eubacteria: Introduction of Characteristics, Living Environments, Ecosystem and Human Relations
References
- Kour D, Khan S.S., et al. "Beneficial fungal communities for sustainable development: Present scenario and future challenges." Journal of Applied Biology & Biotechnology 2023: 1-9 http://www.jabonline.in/
- Raja HA, Miller AN., et al. "Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community." J Nat Prod. 2017 80(3):756-770 https://doi.org/10.1021/acs.jnatprod.6b01085
- Kawahara Y, Oono Y., et al. "Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction." PLoS One. 2012 7(11):e49423 https://doi.org/10.1371/journal.pone.0049423
- Wang H, Wu S., et al. "MicroFisher: Fungal taxonomic classification for metatranscriptomic and metagenomic data using multiple short hypervariable markers." bioRxiv 2024: 23 https://doi.org/10.1101/2024.01.20.576350
- Kokla V, Batrinou A., et al. "Application of imaging spectroscopy, SEM-EDS and DNA analysis for monitoring the preservation status of a manuscript of the first decade of 19th century." Eur. Phys. J. Plus 2023 138: 877 https://doi.org/10.1140/epjp/s13360-023-04502-3
- Paloi S, Mhuantong W., et al. "Using High-Throughput Amplicon Sequencing to Evaluate Intragenomic Variation and Accuracy in Species Identification of Cordyceps Species." J Fungi (Basel). 2021 7(9):767 https://doi.org/10.3390/jof7090767


 Sample Submission Guidelines
Sample Submission Guidelines