Eukaryotic transcription is a key initial step of gene expression, and its precise regulation is important for maintaining cell homeostasis and normal physiological functions of organisms. In recent years, more and more studies show that the disorder of the transcription process is closely related to the occurrence and development of many human diseases. An in-depth analysis of the relationship between eukaryotic transcription abnormalities and diseases from the molecular mechanism level is not only helpful to reveal the pathological process of diseases, but also provides a theoretical basis for the development of targeted therapy strategies.
This paper expounds the relationship between eukaryotic transcription abnormality and diseases, analyzes the pathogenic mechanism of transcription abnormality, introduces its manifestations in diseases, and corresponding treatment strategies.
Brief Introduction of Eukaryotic Transcription
Eukaryotic transcription is the initial core step of gene expression and the key process of transferring DNA genetic information to RNA. In eukaryotic cells, DNA mainly exists on chromosomes in the nucleus, and the transcription process takes place. In this process, one of the double-stranded DNA is used as a template, and with the cooperation of RNA polymerase and various transcription factors, RNA molecules are synthesized according to the principle of base complementary pairing (A-U, T-A, G-C, C-G), thus transferring genetic information from DNA to RNA, laying the foundation for the subsequent protein synthesis.
Eukaryotes have three different types of RNA polymerases, which are responsible for the transcription of genes.
- RNA polymerase I is mainly responsible for the transcription of genes encoding ribosomal RNA (rRNA), which is an important part of ribosomes and participates in the protein synthesis site.
- RNA polymerase II is responsible for the transcription of genes encoding messenger RNA (mRNA) and most non-coding RNA. As a template for protein synthesis, mRNA plays a central role in the translation of gene expression.
- RNA polymerase III mainly transcribes transport RNA (tRNA), 5S rRNA, and some small nuclear RNA (snRNA). tRNA is responsible for transporting amino acids in protein synthesis, 5S rRNA is involved in ribosome assembly, and snRNA is mainly involved in the splicing process of mRNA precursors.
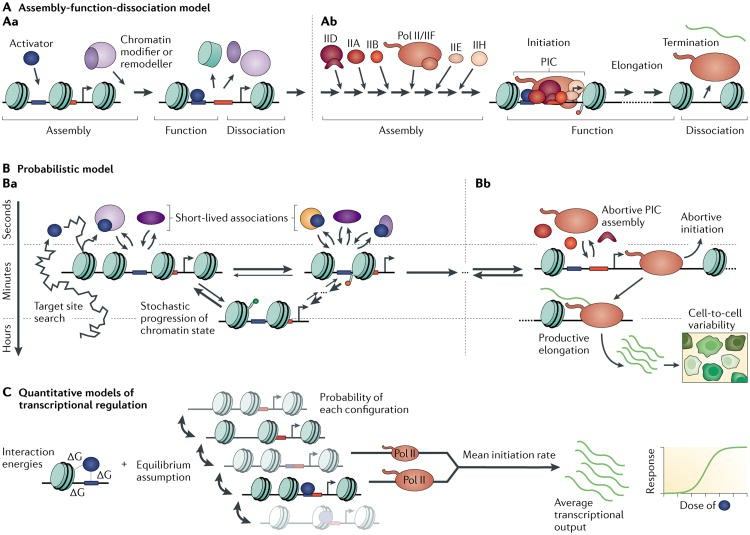
Points of view on transcriptional regulation (Coulon et al., 2013)
Common Eukaryotic Transcription Abnormalities
The precise regulation of the eukaryotic transcription process maintains the normal function of cells. However, influenced by many factors, transcription is prone to abnormality. From the functional imbalance of transcription factors, chromatin structural changes to cis-acting element mutations, these abnormalities interfere with the transmission of genetic information, destroy the balance of gene expression, and then cause cell dysfunction, which has become an important pathological basis for cancer, neurodegenerative diseases, and other diseases.
Transcription Factor Abnormality
Transcription factors are protein molecules that can recognize and bind DNA cis-acting elements, and they play a key role in transcription initiation and regulation. The abnormal transcription factors can be manifested in many forms, such as changes in expression, structural variation, and functional defects. The gene mutation of some transcription factors may lead to a change in their amino acid sequence, which in turn affects the specificity and affinity of DNA binding.
In acute promyelocytic leukemia (APL), PML-RARα fusion protein is produced by chromosome translocation and fusion of the promyelocytic leukemia gene (PML) and retinoic acid receptor α gene (RARα). As an abnormal transcription factor, the fusion protein not only loses the regulatory function of normal transcription factors, but also recruits transcription inhibition complexes, inhibits the transcription of normal hematopoietic-related genes, promotes abnormal cell differentiation and proliferation, and finally causes leukemia.
In addition, the abnormal expression level of transcription factors will also interfere with the normal transcription process. In many cancers, transcription factors encoded by proto-oncogenes such as MYC and NF-κB are often overexpressed. MYC protein can bind to specific E-box sequences on DNA and regulate the transcription of many genes related to cell proliferation, metabolism, and apoptosis. When the MYC gene is abnormally activated or overexpressed, it will lead to uncontrolled cell proliferation and metabolic disorder, and promote the occurrence and development of tumors.
ChIP-seq technology plays an important role in detecting abnormal transcription factors. This technology helps researchers understand the influence of abnormal binding of transcription factors on gene transcription.
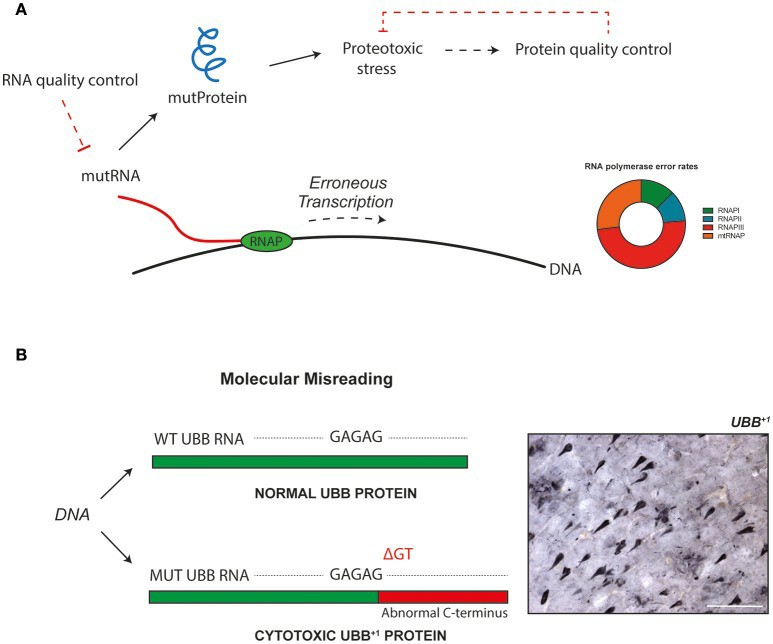
Erroneous gene transcription can bring about abnormal proteins with cytotoxic properties (Verheijen et al., 2017)
Chromatin Structural Change
Chromatin is composed of DNA and histone, and its structural state has a great influence on gene transcription activity. Chromatin structural changes are mainly achieved through histone modification and chromatin remodeling, and abnormal modification and remodeling will lead to transcription disorders. Histone modification includes methylation, acetylation, etc. Histone acetylation is usually related to gene activation, while deacetylation inhibits transcription. In the brain neurons of patients with Alzheimer’s disease, the level of histone acetylation is decreased, which leads to the inhibition of related gene transcription.
Chromatin remodeling complex can change the position and structure of nucleosomes on DNA, and when the complex is dysfunctional, it will affect gene transcription. Hi-C is indispensable for the detection of chromatin structural changes. In addition, ATAC-seq uses the characteristic that transposase can preferentially bind to open chromatin regions, inserts transposons with sequencing linkers into open chromatin regions, and then identifies these regions by high-throughput sequencing.
Cis-acting Element Mutation
Cis-acting elements are specific sequences existing in DNA molecules, including promoters, enhancers, silencers, etc. They regulate the initiation and efficiency of gene transcription by binding to transcription factors. The mutation of these elements will destroy the normal combination of transcription factors and DNA, and then affect gene transcription.
- Mutation of the promoter region may reduce its affinity with RNA polymerase or transcription factor, resulting in a decrease in transcription initiation efficiency. In β-thalassemia, the mutation of the β-globin gene promoter region will reduce the combination of RNA polymerase and promoter, which will significantly reduce the transcription level of the β-globin gene and lead to insufficient synthesis of the β-globin chain, which will eventually lead to anemia symptoms.
- Enhancers are DNA sequence elements that can enhance the transcription activity of genes, and they can play a role far away from promoters. When the enhancer mutates, it may weaken its ability to enhance gene transcription. In some breast cancer cells, the mutation of the enhancer region of proto-oncogene HER2 will affect the combination of related transcription factors, resulting in enhanced transcription of the HER2 gene and over-expression of HER2 protein, which will promote the proliferation and invasion of tumor cells.
- Silencers are DNA sequences that can inhibit gene transcription, and their mutations may cause abnormal activation of genes that should be suppressed. In some hematological tumors, methylation and other modifications or sequence mutations occur in the silencing region of tumor suppressor genes, which will make the silencing function lose, leading to abnormal expression of tumor suppressor genes and fail to play a normal tumor-inhibiting role.
To detect cis-acting element mutations, whole genome sequencing (WGS) and whole-exon sequencing (WES) are widely used. WGS can sequence the whole genome and comprehensively detect DNA sequence variation. WES helps to find the mutation site of cis-acting elements, and clarifies the genetic basis of transcription abnormality.
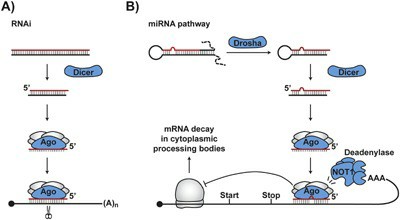
A general outline of Dicer- and Argonaute-dependent post-transcriptional RNA silencing mechanisms in eukaryotes (Hamid et al., 2016)
Diseases Related to Abnormal Eukaryotic Transcription
When the transcription process is abnormal due to various factors, such as transcription factor dysfunction, chromatin structure change, or cis-acting element mutation, the balance of gene expression in cells will be broken. This abnormal transcription is closely related to cancer, neurodegenerative diseases, hereditary diseases and many other diseases, which fundamentally interferes with the normal physiological activities of cells and becomes a key driving factor for the occurrence and development of diseases, and also urges the medical community to explore new strategies for disease treatment based on abnormal transcription.
Cancer
Transcriptional abnormality plays a key role in the development of cancer. Abnormal activation of protooncogenes and inactivation of tumor suppressor genes are often related to abnormal transcription regulation. As an important proto-oncogene, abnormal methylation or over-activation of related transcription factors in the promoter region of the MYC gene will lead to sustained high expression of the MYC gene and promote abnormal cell proliferation, differentiation, and migration.
p53 is an important tumor suppressor gene. When the gene encoding p53 mutates, the transcription factor can’t normally bind to its promoter region, the expression of p53 protein decreases, or its function is lost, and the mechanisms of DNA repair, cycle arrest, and apoptosis of cells are damaged, which eventually leads to tumor occurrence. In addition, abnormal chromatin remodeling is also common in cancer. Some chromatin remodeling complex gene mutations will change the chromatin structure and promote the expression disorder of tumor-related genes.
Neurodegenerative Diseases
Neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) are also closely related to abnormal eukaryotic transcription. In the brain neurons of AD patients, the transcription regulation of β-amyloid precursor protein (APP) gene is abnormal, which leads to the over-expression of APP, and then produces too much β-amyloid, forming senile plaques, causing neuronal damage and death. At the same time, the abnormal modification of the Tau protein gene after transcription makes Tau protein hyperphosphorylated and forms neurofibrillary tangles, which affects the normal function of neurons. In PD, the transcription regulation of the α-synuclein gene is out of balance, which leads to the abnormal aggregation of α-synuclein and the formation of Lewy bodies, which damage dopaminergic neurons and cause motor dysfunction.
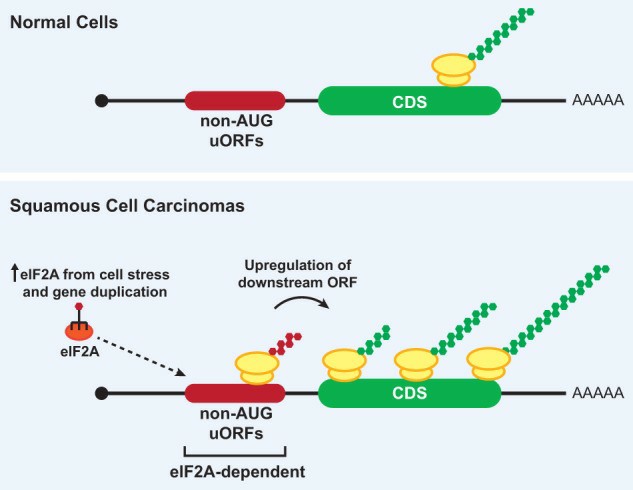
eIF2A regulates translation of non-AUG uORFs in SCCs (Kearse et al., 2017)
Genetic Diseases
Many hereditary diseases are caused by abnormal gene transcription. For example, thalassemia is caused by abnormal transcription regulation of globin gene, which leads to the reduction or deletion of globin chain synthesis and affects the normal assembly and function of hemoglobin; Cystic fibrosis is caused by abnormal transcription of cystic fibrosis transmembrane conductance regulator (CFTR) gene, which leads to insufficient expression or functional defect of CFTR protein, which leads to ion transport disorder in cell membrane and leads to multiple organ diseases such as lung and pancreas.
Treatments of Diseases Based on Transcription Abnormal
With the understanding of transcription regulation networks, strategies such as small molecule drugs, gene therapy, and epigenetic regulation therapy are constantly emerging. Targeted intervention in the abnormal transcription process provides diversified intervention means for disease treatment.
Small Molecule Drug
It is an important therapeutic strategy to develop small-molecular drugs that can target and regulate transcription abnormalities. Given abnormal transcription factors, small molecular compounds can inhibit the activity of abnormally activated transcription factors or promote the normal expression and functional recovery of transcription factors. Given some transcription factors that are over-activated in cancer, specific inhibitors are developed to block their combination with DNA, thus inhibiting the transcription of tumor-related genes. In addition, small molecule drugs can also be used to regulate histone modification, such as histone deacetylase (HDAC) inhibitors, which can increase histone acetylation level, change chromatin structure, and restore normal transcription of some genes by inhibiting HDAC activity.
Gene Therapy
Gene therapy aims at correcting or compensating for abnormal gene transcription. The normal gene is introduced into the patient’s cells through the vector to replace or repair the mutant gene and restore normal transcription. Adeno-associated virus (AAV) vector was used to introduce the normal CFTR gene into cystic fibrosis patients’ cells, so that they could normally transcribe and express CFTR protein and improve the ion transport function of the cell membrane. In addition, RNA interference (RNAi) technology can be used to design small interfering RNA (siRNA) or short hairpin RNA (shRNA) for abnormal transcription-related genes, which can specifically inhibit the transcription and expression of abnormal genes, showing potential application value in the treatment of cancer and some genetic diseases.
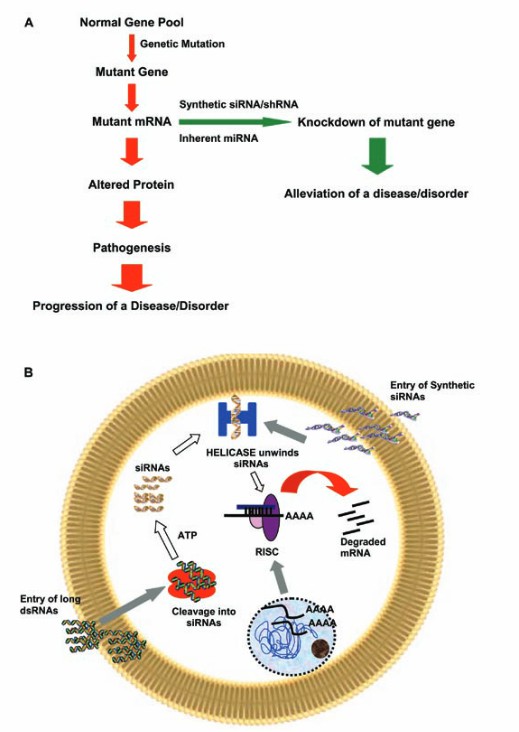
Mechanism of RNAi (Pushparaj et al., 2008)
Epigenetic Regulation Therapy
Given the abnormal chromatin structure and epigenetic modification, epigenetic regulation drugs can be developed. In addition to the HDAC inhibitors mentioned above, DNA methyltransferase (DNMT) inhibitors can reduce DNA methylation and reactivate silent genes. In some hematological tumors, DNMT inhibitors can restore the normal expression of tumor suppressor genes and inhibit the growth of tumor cells. In addition, by regulating the expression and function of non-coding RNA (such as miRNA and lncRNA), it is also one of the research directions of epigenetic regulation therapy to interfere with the transcription regulation network.
Conclusion
Eukaryotic transcription abnormality is closely related to the occurrence and development of many diseases, from transcription factor abnormality, chromatin structure change, to cis-acting element mutation and other factors, which together lead to the imbalance of gene transcription regulation, leading to a series of diseases such as cancer, neurodegenerative diseases, and hereditary diseases. Sequencing technology plays an indispensable role in exploring the mechanism of eukaryotic transcription abnormality. RNA-seq can comprehensively detect all transcripts in cells, it can not only quantify the expression level of mRNA, but also find the abnormal expression of non-coding RNA, thus revealing the overall changes of transcriptome in disease state.
With the deepening of the research on the mechanism of eukaryotic transcription abnormality, the treatment methods of diseases based on transcription abnormality are gradually diversified, and small molecule drugs, gene therapy, and epigenetic regulation therapy bring new hope for the treatment of related diseases.
Learn more information, you can read:
References
- Kornberg RD. "The molecular basis of eukaryotic transcription." Proc Natl Acad Sci U S A. 2007 104(32):12955-61 https://doi.org/10.1073/pnas.0704138104
- Verheijen BM, van Leeuwen FW. "Commentary: The landscape of transcription errors in eukaryotic cells." Front Genet. 2017 8:219 https://doi.org/10.3389/fgene.2017.00219
- Hamid FM, Makeyev EV. "Exaptive origins of regulated mRNA decay in eukaryotes." Bioessays. 2016 38(9):830-8 https://doi.org/10.1002/bies.201600100
- Kearse MG, Wilusz JE. "Non-AUG translation: a new start for protein synthesis in eukaryotes." Genes Dev. 2017 31(17):1717-1731 https://doi.org/10.1101/gad.305250.117
- Pushparaj PN, Aarthi JJ., et al. "siRNA, miRNA, and shRNA: in vivo applications." J Dent Res. 2008 (11): 992-1003 https://doi.org/10.1177/154405910808701109
- Coulon A, Chow CC., et al. "Eukaryotic transcriptional dynamics: from single molecules to cell populations." Nat Rev Genet. 2013 14(8): 572-84 https://doi.org/10.1038/nrg3484


 Sample Submission Guidelines
Sample Submission Guidelines