In the evolution of biotechnology applications, DNA extraction, a fundamental technology for analyzing genetic information, has consistently occupied a core and irreplaceable position. From the molecular verification of Mendel’s genetic factor hypothesis to the vast practice of the Human Genome Project, the acquisition of high-quality DNA constitutes the logical starting point for scientific discovery and technological transformation.
This technology not only needs to break through the physical barriers and biochemical interference of different sample types but also needs to achieve a precise balance between the integrity, purity, and yield of nucleic acid molecules. The innovation of its methodology and the construction of standardization system continue to promote the leap-forward development of frontier fields such as genomics, precision medicine, and synthetic biology.
This paper comprehensively introduces the progress of DNA extraction technology, and shows the key role and development direction of this technology in biological research and practical application.
What is the Need to Extract DNA
As the core carrier of genetic information of living organisms, the importance of DNA extraction technology plays a key role in modern biological research and application. In the field of basic scientific research, there are strict requirements for obtaining high-purity DNA samples in frontier fields such as gene structure and function analysis, whole genome sequencing analysis, and gene expression profile research.
DNA extraction is the first step in genomics research. By extracting genomic DNA from cells of different species, scientists can draw the genome map of species and analyze the arrangement of genes and the distribution of regulatory elements.
- Gene expression analysis depends on DNA pollution control before RNA extraction and DNA extraction in promoter region is the key to studying transcription regulation. Chromatin immunoprecipitation sequencing (ChIP-seq) technology needs to extract DNA from cross-linked chromatin in order to identify the genomic regions bound to transcription factors.
- DNA extraction from tumor tissues is the basis of cancer genomics research. By extracting DNA from the tumor and normal tissues adjacent to the tumor and sequencing the whole genome, it was found that there were a large number of somatic mutations in tumor cells. The discovery of these mutations not only reveals the molecular pathway of tumorigenesis but also provides a target for targeted drug development.

Sensing and responding to damaged DNA (Jackson et al., 2018)
- DNA extraction is a prerequisite technology for molecular marker-assisted breeding (MAS). In rice breeding for rice blast resistance, extracting DNA from different strains and detecting molecular markers closely linked with disease-resistant genes can accelerate the screening of disease-resistant varieties and shorten the traditional breeding cycle from 8-10 years to 4-5 years.
- The supervision and safety evaluation of genetically modified crops depends on DNA extraction technology. By extracting the DNA of genetically modified crops, qualitative PCR and quantitative PCR techniques can be used to detect the integration and copy number of foreign genes (such as insect-resistant gene Bt) and ensure that genetically modified crops meet the safety standards.
- In animal husbandry, DNA extraction is used for breeding excellent breeds of livestock and poultry. Genome-wide selection (GS) is carried out by extracting breeding stock DNA, and individual breeding potential is evaluated by genome-estimated breeding value (GEBV), which can improve selection accuracy and accelerate genetic progress.
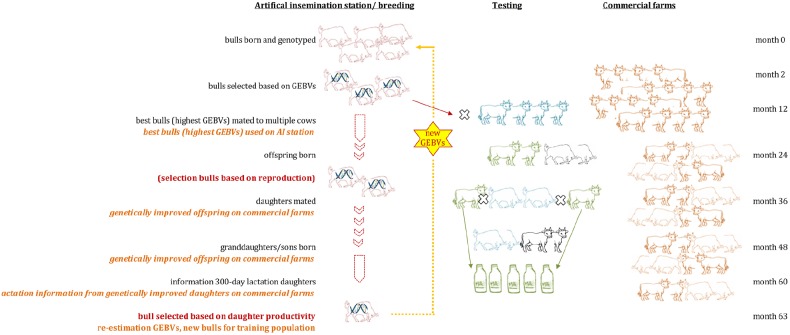
Marker enhanced breeding using Genomic Selection in dairy cattle (Jonas et al., 2015)
Strategies for DNA Extraction from Different Samples
The physical and chemical properties and components of different biological samples are significantly different, which makes it necessary to adopt targeted strategies for DNA extraction. Animal tissues contain protein and nuclease, plants are rich in polysaccharides and polyphenols, and microorganisms have unique cell wall structure. Paleontological samples are faced with DNA degradation and pollution, so it is necessary to optimize the cracking, purification and anti-degradation scheme according to the characteristics of the samples.
DNA Extraction Strategy of Animal Tissue Samples
A. Blood sample
Blood sample is the most commonly used sample type in medical and biological research, and its DNA mainly exists in white blood cells. The core of the extraction strategy is to efficiently remove red blood cells and lyse white blood cells. For fresh blood, EDTA or sodium citrate is usually used for anticoagulation to avoid DNA degradation caused by coagulation. Classical blood DNA extraction methods include:
Phenol-chloroform extraction method: This method is based on the difference in distribution coefficients of different substances in the organic phase and water phase.
- First, most of the red blood cells were removed by red blood cell lysate, and then the white blood cells were lysed by adding a buffer containing protease K to release DNA.
- Then it was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) to denature the protein and distribute it to the organic phase, while the DNA remained in the water phase.
- Finally, pure DNA was obtained by ethanol precipitation. The DNA extracted by this method has high purity, but the operation is complicated, and phenolic reagents are toxic.
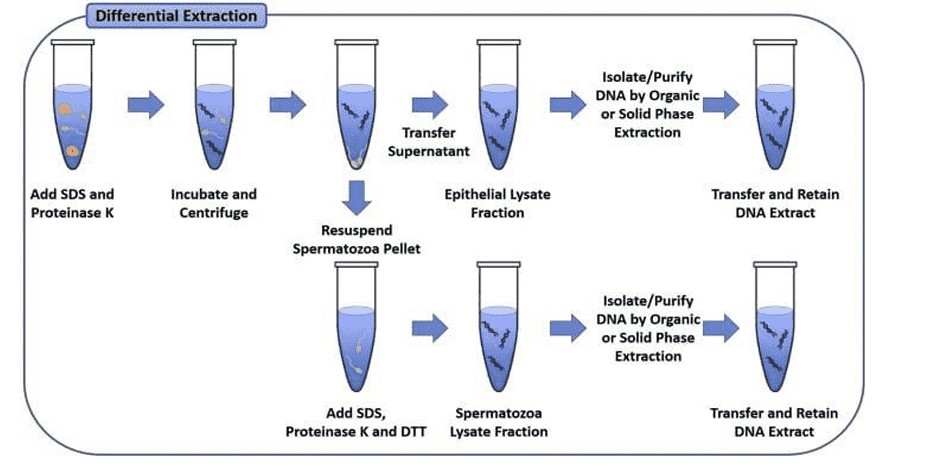
SDS-Proteinase K DNA extraction method (Komal et al., 2023)
Silica gel membrane column centrifugation: Using the specific adsorption characteristics of silica gel membrane to DNA. Under the condition of high salt and low pH, DNA binds to the silica gel membrane, while impurities such as protein and polysaccharides flow out with the liquid. After washing many times to remove residual impurities, DNA fell off the silica gel membrane under the action of elution buffer with low salt and high pH, and rapid separation was realized. This method is simple and time-consuming, suitable for Qualcomm sample processing, and widely used in clinical diagnosis and scientific research laboratories.
B. Tissue sample
The difficulty of DNA extraction from animal tissues (such as liver, muscle, and tumor) lies in the effective lysis of tissue cells and the inhibition of nuclease. Fresh or frozen tissue (stored at -80℃) should be cut or ground first to destroy the tissue matrix:
- Freeze grinding method: put the tissue in liquid nitrogen and grind it into powder to avoid DNA degradation caused by repeated freezing and thawing, which is suitable for samples rich in connective tissue.
- Protease K digestion method: The commonly used lysate formula is 10mM Tris-Cl (pH 8.0), 100 mM EDTA, 0.5% SDS, and 200μg/mL protease K, and incubated at 37-55℃ for several hours until the tissue is completely dissolved. For formalin-fixed paraffin-embedded (FFPE) tissues, it is necessary to dewax (soak in xylene for 2-3 times), then digest with protease K and cross-link at high temperature (for example, incubate at 65℃ for 2 hours), but the extracted DNA is often fragmented and suitable for short-segment PCR.
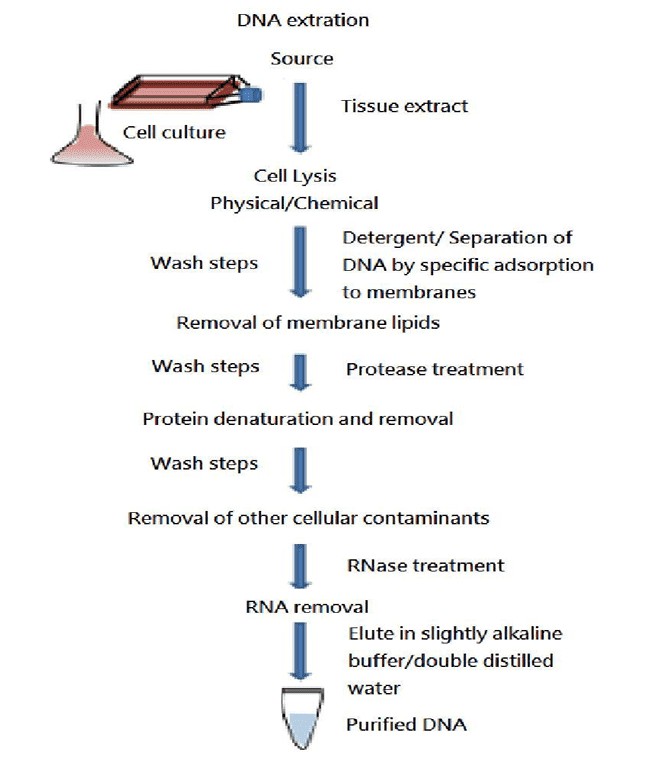
Basic steps in DNA extraction (Komal et al., 2023)
DNA Extraction from Plant Tissue Sample
A. Leaves and tender tissues
Plant cells have hard cell walls and are rich in secondary metabolites such as polysaccharides and polyphenols, which are easy to combine with DNA to form viscous complexes, which affect the extraction purity. The classical cetyltrimethylammonium bromide (CTAB) method is the gold standard for plant DNA extraction. The core step of the CTAB method:
- Grinding leaves into fine powder in liquid nitrogen, adding preheated CTAB buffer (containing 2% CTAB, 1.4M NaCl, 100mM Tris-Cl, 20mM EDTA and 0.2% β-mercaptoethanol), and lysing cells in a water bath at 65℃ for 30-60min.
- Protein was removed by chloroform-isoamyl alcohol (24:1) extraction, and DNA was precipitated by adding 1/10 volume of 5M NaCl and equal volume of isopropanol. After washing with 70% ethanol, it was dissolved in TE buffer.
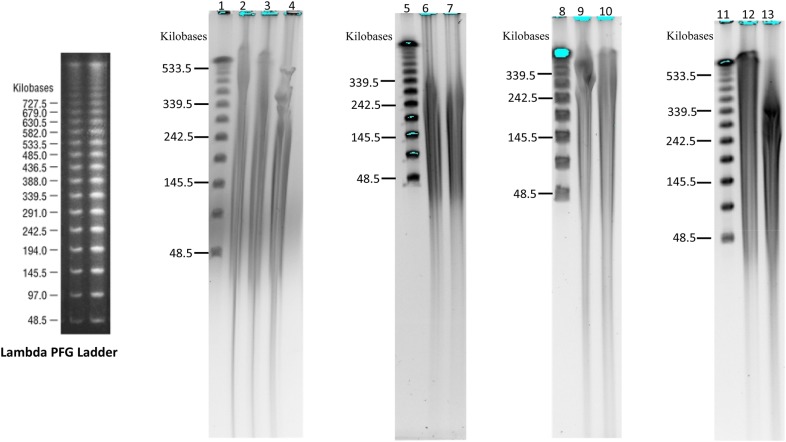
The high-molecular-weight genomic DNA fractionation by Pulsed-field gel system (Li et al., 2020)
Optimization strategy: For samples rich in polyphenols (such as tea and grapes), 2-5% PVP (polyvinylpyrrolidone) can be added to the pyrolysis liquid to reduce oxidation by using its characteristics of adsorbing polyphenols. High salt concentration (1.4M NaCl) can inhibit the co-precipitation of polysaccharide and DNA.
B. Seeds and storage organs
Seeds, tubers, tubers, etc. are rich in starch and storage protein, so it is necessary to strengthen impurity removal during extraction:
- High-salt-low-pH method: Buffer containing 2M NaCl and 0.1M potassium acetate is used to reduce the solubility of DNA in high-salt solution and promote starch precipitation. Acidic condition (pH 4.8) can inhibit the activity of DNase and reduce protein binding.
- PEG precipitation method: 10-20% PEG 6000 and 0.5M NaCl were added to the DNA extract, and the DNA was selectively precipitated by the precipitation of macromolecules by PEG, while starch and micromolecule sugar remained in the supernatant.
Corn seeds, potato tubers, and other samples need to remove starch particles first, and starch can be precipitated by differential centrifugation (2000g centrifugation for 10 minutes), and then DNA extraction is carried out on the supernatant.
Optimization of DNA Extraction Technology
With the development of life science research towards precision and Qualcomm quantification, the limitations of traditional DNA extraction technology in efficiency, purity and applicability are increasingly prominent. From the toxic hidden danger of phenol-chloroform extraction to the cracking problem of complex samples, from the loss risk of trace DNA to the demand upgrade of automatic processing, technology optimization has become the key path to breaking through the research bottleneck.
Extraction Method Improvement
Although the traditional phenol-chloroform extraction method has high extraction efficiency, it has some shortcomings such as toxic reagents and complicated operation. With the development of technology, a variety of new DNA extraction methods came into being. The Silica gel column adsorption method is one of the most widely used methods at present. Its principle is to use a silica gel membrane to adsorb DNA under high salt conditions and release DNA under low salt conditions.
This method is simple and time-consuming and is suitable for automatic Qualcomm extraction. The magnetic bead method is used to extract DNA by combining functional groups modified on the surface of magnetic nanoparticles with DNA and separating DNA under the action of the external magnetic field. This method has the advantages of high automation and good biological safety and is especially suitable for large-scale detection of clinical samples.
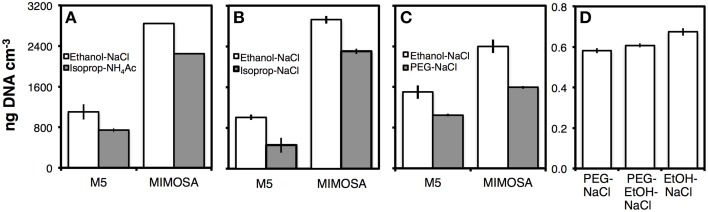
Comparisons of DNA yields by different precipitation methods in relation to Ethanol-NaCl precipitation (Lever et al., 2015)
Automation and Qualcomm Quantity Technology
With the development of genomics research, higher requirements are put forward for the throughput and efficiency of DNA extraction. By integrating the steps of sample processing, reagent addition, and nucleic acid separation, the automatic DNA extractor realizes the automatic operation of DNA extraction, reduces human error, and improves extraction efficiency. At present, there are many automatic extraction platforms on the market:
- The 96-well-plate Qualcomm extractor, which can process dozens to hundreds of samples at the same time and is suitable for large-scale epidemiological investigation, genome sequencing, and other projects.
- Microfluidic chip technology has also been applied to DNA extraction. By constructing microchannels and reaction units on the chip, rapid DNA extraction of trace samples is realized, which has the characteristics of small volume, less sample consumption, and simple operation.
Common Problems and Solutions of DNA Extraction
In the process of DNA extraction, various problems often occur due to factors such as sample characteristics and operating specifications. Such as low extraction concentration, insufficient purity, DNA degradation, and the existence of inhibitor, these problems will affect the follow-up experiments.
Low DNA Concentration
Low DNA concentration is a common problem in DNA extraction, which may be caused by insufficient initial sample amount, insufficient cell lysis, and low efficiency of DNA adsorption or elution. To solve this problem, we should first ensure that the initial amount of samples is sufficient.
- For blood samples, it is recommended to use at least 200 μL of anticoagulant whole blood.
- For tissue samples, fresh or correctly preserved tissues should be selected to avoid repeated freezing and thawing. Cell lysis is a key step.
- For samples that are difficult to lyse, such as gram-positive bacteria and plant cells, various lysis methods can be used, such as enzymolysis and mechanical crushing.
In the process of DNA adsorption, we should pay attention to the salt concentration and pH value of the adsorption buffer to ensure that DNA is fully adsorbed on silica gel membrane or magnetic beads. When eluting, the elution efficiency of DNA can be improved by using a preheated elution buffer (such as TE buffer or water) and prolonging the elution time appropriately.
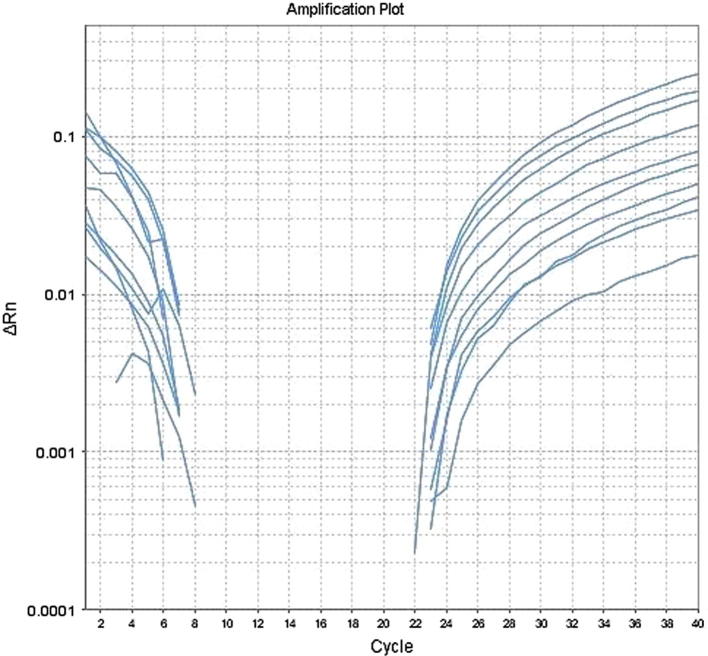
Amplification plot for the gene CYP2C9 exhibits the integrity of DNA samples extracted from whole blood (Qamar et al., 2017)
Low DNA Purity
The low purity of DNA usually shows that the ratio of A260/A280 deviates from 1.8 (the ratio of pure DNA is 1.8), or the ratio of A260/A230 is too low (normally it should be greater than 2.0), suggesting that there are impurities such as protein, phenol, and polysaccharide. Protein pollution may be due to insufficient cell lysis or insufficient protease K activity. Therefore, the dosage of protease K can be increased or the digestion time can be prolonged, and at the same time, sufficient oscillation can be ensured during phenol-chloroform extraction, so that protein can be fully denatured and precipitated.
Phenol residue can lead to an increase in the A280 ratio, which can be solved by increasing chloroform extraction times or using a phenol removal column. Polysaccharide pollution is common in plant samples. High concentration NaCl or PEG can be added to the extraction buffer to precipitate polysaccharide impurities or ethanol fractionation can be used to separate DNA and polysaccharide by using the solubility difference of different concentrations of ethanol.
DNA Degradation
DNA degradation is characterized by band dispersion or smear in electrophoresis detection, which is mainly caused by nuclease pollution, improper sample preservation, or violent operation during extraction. Preventing nuclease pollution is the key. RNase-free/DNase-free reagents and consumables should be used during the experiment, and operators should wear gloves to avoid skin contact with samples. Sample preservation should adopt correct methods, such as using anticoagulant (EDTA or sodium citrate) for blood samples, and immediately freezing tissue samples at-80℃ to avoid repeated freezing and thawing.
Violent vibration and repeated sucking should be avoided in the extraction process, especially for the extraction of high molecular weight DNA, such as genomic DNA, mild cracking and separation methods can be adopted, such as low-speed centrifugation, gentle inversion, and mixing.
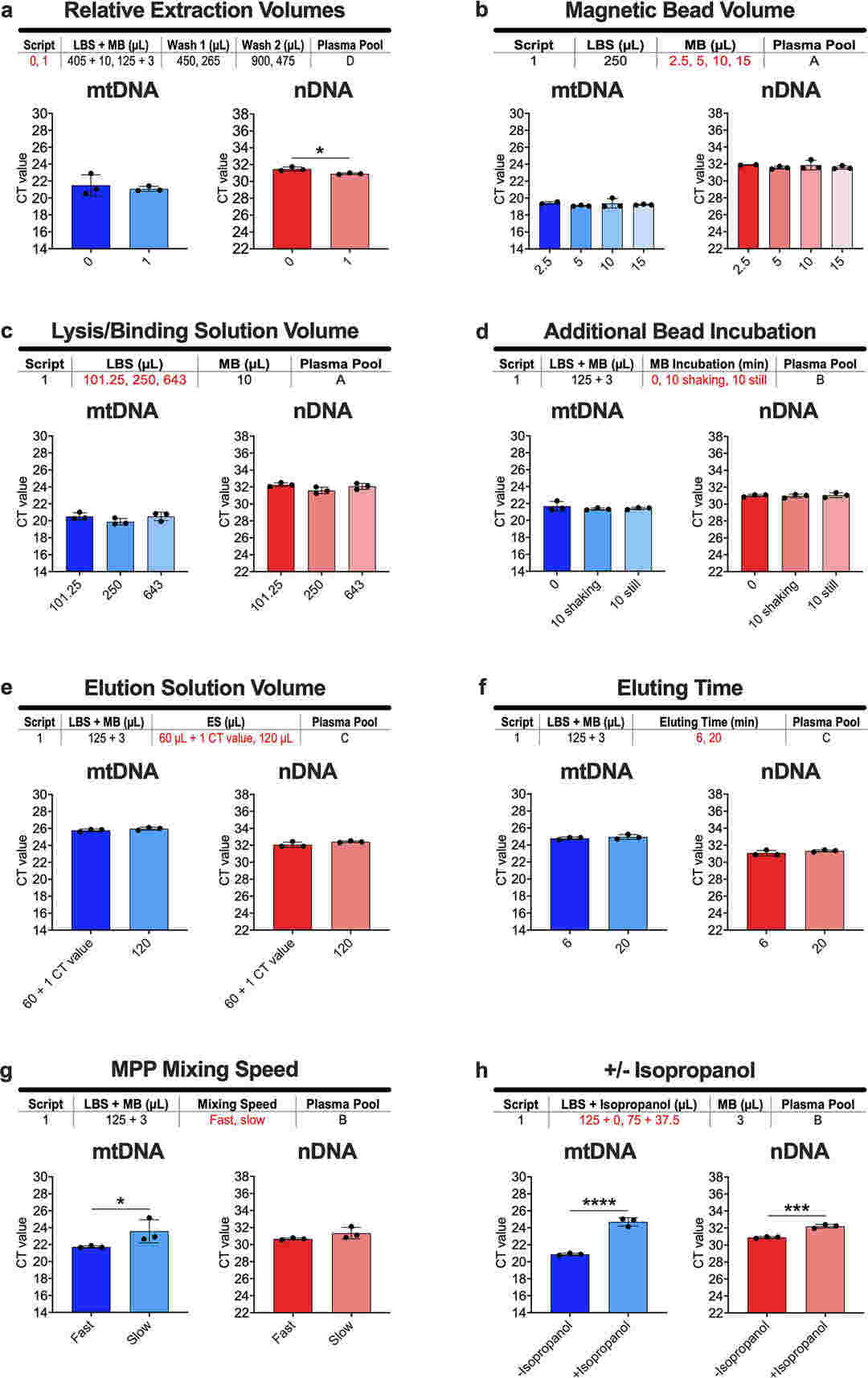
Optimization of extraction parameters (Ware et al., 2020)
Conclusion
The development of DNA extraction technology has always been closely linked with the demand for life science research and application. From the initial phenol-chloroform extraction method to the automatic Qualcomm extraction method, from a single type of sample to a complex environmental sample, DNA extraction technology has made remarkable progress in methods, efficiency, and applicability. However, in the face of samples from different sources and different preservation States, DNA extraction still faces many challenges, such as efficient cracking of special samples, enrichment of trace samples, and removal of complex impurities.
In the future, the development of DNA extraction technology will move towards a more efficient, automated, and intelligent direction. The combination of new nano-materials, microfluidic technology, and artificial intelligence is expected to realize the full automation and intelligent operation of DNA extraction, improve the efficiency and quality of extraction, and lower the operating threshold.
Learn More
References:
- Jackson M, Marks L, May GHW, Wilson JB. "The genetic basis of disease." Essays Biochem. 2018 62(5): 643-723 https://doi.org/10.1042/ebc20170053
- Jonas E, de Koning DJ. "Genomic selection needs to be carefully assessed to meet specific requirements in livestock breeding programs." Front Genet. 2015 6: 49 https://doi.org/10.3389/fgene.2015.00049
- Li Z, Parris S, Saski CA. "A simple plant high-molecular-weight DNA extraction method suitable for single-molecule technologies." Plant Methods. 2020 16: 38 https://doi.org/10.1186/s13007-020-00579-4
- Lever MA, Torti A., et al. "A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types." Front Microbiol. 2015 6: 476 https://doi.org/10.3389/fmicb.2015.00476
- Qamar W, Khan MR, Arafah A. "Optimization of conditions to extract high quality DNA for PCR analysis from whole blood using SDS-proteinase K method." Saudi J Biol Sci. 2017 24(7): 1465-1469 https://doi.org/10.1016/j.sjbs.2016.09.016
- Ware SA, Desai N., et al. "An automated, high-throughput methodology optimized for quantitative cell-free mitochondrial and nuclear DNA isolation from plasma." J Biol Chem. 2020 Nov 13;295(46):15677-15691 https://doi.org/10.1074/jbc.ra120.015237


 Sample Submission Guidelines
Sample Submission Guidelines