Understanding microbial diversity is no longer optional—it has become a cornerstone of modern research across environmental science, agriculture, and biomedical fields. Microbes shape ecosystems, drive nutrient cycles, and even influence host health. However, researchers often face challenges such as complex sample types, limited budgets, and different scientific objectives. These hurdles make choosing the right sequencing approach crucial for achieving meaningful results.
At CD Genomics, we provide a comprehensive portfolio of microbial diversity analysis services that range from standard amplicon-based methods to advanced whole-genome sequencing solutions. Whether your project focuses on exploring microbial community structure, tracking ecological changes, or uncovering functional genes, our services deliver precise, reliable, and publication-ready data.
In this article, we provide a complete guide to microbial diversity analysis methods—from conventional 16S/18S/ITS amplicon sequencing to cutting-edge metagenomics, full-length sequencing, eDNA approaches, and absolute quantification. By comparing their principles, advantages, and applications, we aim to help you make informed choices for your microbial research projects.

16S/18S/ITS Sequencing
A Classic Starting Point for Microbial Studies
When researchers first begin exploring microbial communities, they often turn to amplicon sequencing. This method targets highly conserved regions of microbial genomes—such as the 16S rRNA gene in bacteria, the 18S rRNA gene in eukaryotes, or the ITS region in fungi. By amplifying and sequencing these regions, scientists can quickly determine "who is there"in a given sample.
How It Works in Practice
The workflow is straightforward:
- PCR amplification of the target region
- Library construction for sequencing
- High-throughput sequencing on Illumina platforms
- Bioinformatics analysis for OTU/ASV clustering, taxonomic classification, and diversity metrics
This pipeline is well-established, cost-effective, and supported by extensive reference databases—making it the most widely used method in microbial ecology.
Why Researchers Choose Amplicon Sequencing
- Cost-Effective & High Throughput: Enables affordable profiling across large sample sets.
- Mature & Reliable: Supported by decades of published data, ensuring comparability.
- Broad Coverage: Suitable for bacteria, fungi, archaea, and eukaryotes.
Research Scenarios Where It Excels
Amplicon sequencing remains a powerful tool for:
- Initial Community Profiling: A quick way to screen microbial structure before moving into deeper metagenomics.
- Ecological & Microbiota Monitoring: Tracking changes in soil, water, or host-associated microbiomes.
- Long-Term Community Surveys: Building baselines for environmental and agricultural studies.
Takeaway for Researchers:
If your primary goal is to obtain a fast and economical snapshot of community composition, 16S/18S/ITS sequencing remains the most practical entry point. For deeper resolution or functional insights, you may consider full-length sequencing or metagenomics—but as a starting point, this method provides a solid foundation for almost any microbial diversity study.
Metagenomic Sequencing
A Comprehensive View of Microbial Communities
While amplicon sequencing tells us who is present in a microbial community, metagenomic sequencing takes it a step further. By directly sequencing all genomic DNA in a sample, researchers gain insights not only into community composition but also into the functional potential of microbes—the genes they carry, the pathways they drive, and even their resistance elements.
How Metagenomics Works
- Extract total DNA from the sample
- Construct sequencing libraries
- Sequence using Illumina, PacBio, or Nanopore platforms
- Perform rigorous quality control and host DNA removal
- Assemble genomes, annotate genes, and profile community structure and function
This method eliminates the amplification bias of PCR, capturing a more accurate and holistic picture of microbial ecosystems.
Why Researchers Choose Metagenomics
- Unbiased Community Detection: No need for primers, so rare and unexpected organisms can be detected.
- Functional Profiling: Identifies resistance genes, metabolic pathways, and virulence factors.
- Broad Coverage: Detects bacteria, archaea, fungi, viruses, and protists in a single run.
- Supports Genome Assembly: Enables reconstruction of near-complete microbial genomes.
Research Scenarios Where It Excels
- Functional and Pathway Analysis: Mapping microbial contributions to metabolism and ecology.
- Resistance Gene Detection: Critical for tracking the spread of antimicrobial resistance.
- Low-Abundance Microbe Studies: Ideal for soil, water, and host-associated samples with complex microbial signatures.
- Source Tracking: Helps link microbes back to their environmental or host origins.
Takeaway for Researchers
If your research questions go beyond "who is there" and move toward "what are they doing," metagenomic sequencing is the most powerful choice. It requires more data and deeper analysis compared to amplicon methods, but it offers a level of detail—both taxonomic and functional—that no other approach can match.
2bRAD-M
Bridging the Gap Between Amplicon and Metagenomics
For researchers who need higher resolution than amplicon sequencing but want to avoid the cost and data burden of full metagenomics, 2bRAD-M (Type II-B Restriction Site Associated DNA Sequencing ) offers a smart alternative. This technique uses type II-B restriction enzymes to cut DNA at specific recognition sites, generating uniform short fragments that represent the microbial genome without PCR amplification.
How 2bRAD-M Works
- Restriction enzyme digestion of microbial DNA
- Ligation of sequencing adapters to digested fragments
- Library construction and sequencing
- Alignment to reference databases for species and subspecies classification
- Quantitative abundance profiling
Because it bypasses PCR, 2bRAD-M reduces amplification bias while still keeping data volumes manageable compared to shotgun sequencing.
 Schematic workflow of human thanatomicrobiome analysis using 16S rRNA, metagenomics, and 2bRAD-M sequencing. (Huang, X., et al., Sci Data 2024).
Schematic workflow of human thanatomicrobiome analysis using 16S rRNA, metagenomics, and 2bRAD-M sequencing. (Huang, X., et al., Sci Data 2024).
Why Researchers Choose 2bRAD-M
- No PCR Required: Ensures more accurate representation of microbial communities.
- High Taxonomic Resolution: Identifies microbes at species and even subspecies level.
- Cost-Effective: Requires less sequencing data than metagenomics, lowering expenses.
- Fast Library Preparation: Streamlined workflow accelerates project timelines.
Research Scenarios Where It Excels
- Large-Scale Microbial Profiling: Ideal for population studies with many samples.
- Detecting Subtle Variations: Useful when distinguishing closely related microbial strains.
- Budget-Conscious Projects: Provides species-level resolution without the cost of deep metagenomics.
Takeaway for Researchers
If your project requires species-level precision but your resources are limited, 2bRAD-M provides the perfect balance between resolution, cost, and efficiency. It is particularly valuable for large-scale studies where both accuracy and affordability matter.
Full-Length 16S/18S/ITS Sequencing
Although short-read amplicon sequencing is effective for community profiling, it often lacks the resolution needed to distinguish closely related species. Full-length 16S/18S/ITS sequencing, powered by long-read platforms such as PacBio, overcomes this limitation by capturing the entire marker gene—V1 to V9 for 16S rRNA, full 18S for eukaryotes, and the complete ITS region for fungi.
How Full-Length Sequencing Works
- PCR amplification of full-length ribosomal or ITS regions
- Long-read library preparation
- Sequencing on PacBio platforms
- Data quality filtering and error correction
- ASV/OTU analysis and species-level classification
This extended coverage dramatically improves taxonomic accuracy, providing researchers with reliable insights at species or even strain level.
Why Researchers Choose Full-Length Sequencing
- High-Resolution Taxonomy: Achieves more precise classification compared to short-read approaches.
- Improved Accuracy: Reduces misclassification errors common in partial-region sequencing.
- Targeted Species Detection: Enables focused studies of key microbes in complex communities.
Research Scenarios Where It Excels
- High-Precision Community Analysis: For projects that demand species-level annotation.
- Agriculture and Fermentation Studies: Identifies functional microbes critical to crop health or fermentation quality.
- Phylogenetic and Evolutionary Research: Provides longer reads for constructing reliable evolutionary relationships.
Takeaway for Researchers
If your research depends on pinpointing microbes with high confidence, full-length 16S/18S/ITS sequencing is the method of choice. It combines the accessibility of amplicon sequencing with the accuracy of long-read technologies, giving you a sharper lens to study microbial communities.
Absolute Quantification 16S/18S/ITS Sequencing
Overcoming the Limits of Relative Data
Most conventional diversity methods, including amplicon sequencing, report results in terms of relative abundance. While this shows community composition, it does not reveal the true number of microbes in a given environment. Absolute quantification sequencing solves this problem by introducing synthetic spike-in standards into the workflow, enabling direct measurement of microbial copy numbers per unit mass or volume.
How Absolute Quantification Works
- Sample preparation with spike-in standard addition
- PCR amplification of 16S, 18S, or ITS regions
- Library construction and high-throughput sequencing
- Data modeling with internal standards for absolute quantification
- Conversion of sequencing reads into true microbial counts
This approach transforms microbial profiling from a comparative tool into a quantitative measurement system.
Why Researchers Choose Absolute Quantification
- True Quantitative Accuracy: Provides microbial counts rather than relative percentages.
- Dynamic Community Profiling: Captures time-series changes with precision.
- Improved Ecological Interpretation: Allows cross-sample and cross-condition comparisons on an absolute scale.
Research Scenarios Where It Excels
- Longitudinal Microbiome Studies: Tracking microbial dynamics across time points.
- Microbial Load Assessment: Determining total biomass in soil, water, or host samples.
- Ecological Modeling: Supporting systems biology studies where quantitative parameters are required.
Takeaway for Researchers
If your study requires absolute microbial counts to build ecological models, monitor time-dependent dynamics, or compare across diverse conditions, absolute quantification sequencing provides the rigor you need. It is particularly powerful for turning descriptive microbiome data into predictive ecological insights.
Environmental DNA (eDNA) Analysis
A Non-Invasive Window Into Ecosystems
Instead of directly sampling organisms, environmental DNA (eDNA) analysis allows researchers to study biodiversity by extracting free-floating DNA from soil, water, or air. This technique has transformed ecological monitoring, enabling scientists to detect a wide range of organisms—including plants, animals, and microbes—without disturbing the environment.
How eDNA Analysis Works
- Collect environmental samples (e.g., water, soil, sediment, or air filters)
- Extract total DNA from the sample
- Amplify target regions such as COI, 12S, 16S, or trnL genes
- Perform high-throughput sequencing
- Align and annotate sequences to identify organisms present
By capturing DNA traces shed into the environment, eDNA makes it possible to reconstruct an ecosystem's biodiversity profile with remarkable breadth.
Why Researchers Choose eDNA
- Non-Invasive Sampling: Ideal for sensitive or protected habitats.
- Broad Taxonomic Detection: Detects plants, animals, fungi, and microbes simultaneously.
- Supports Long-Term Monitoring: Easily adapted to seasonal or longitudinal studies.
Research Scenarios Where It Excels
- Aquatic and Soil Surveys: Profiling biodiversity in lakes, rivers, oceans, and terrestrial ecosystems.
- Rare or Invasive Species Detection: Identifying elusive, endangered, or invasive species from trace DNA.
- Conservation and Resource Management: Supporting habitat protection, restoration, and ecological planning.
 Proportion of the sequencing output allocated to the different species. (Díaz, C. et al. Sci Rep 2020)
Proportion of the sequencing output allocated to the different species. (Díaz, C. et al. Sci Rep 2020)
Takeaway for Researchers
If your goal is ecosystem-level biodiversity monitoring with minimal impact on the environment, eDNA is the method of choice. It combines non-invasive sampling with broad taxonomic coverage, making it a powerful tool for conservation biology, ecology, and environmental monitoring.
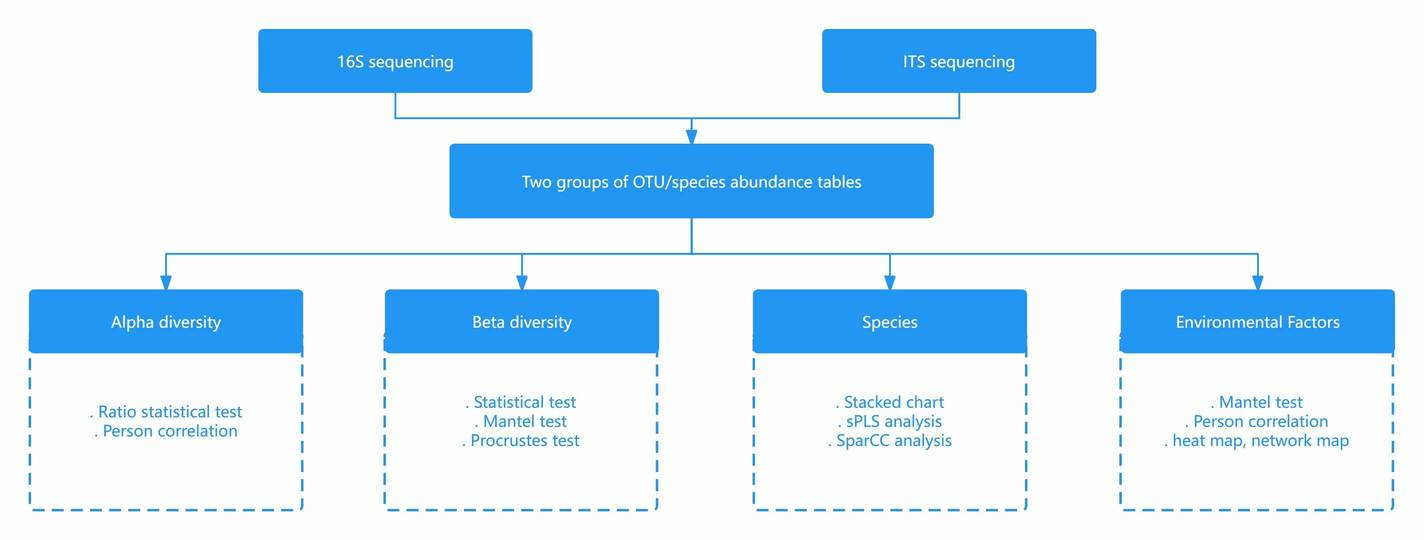
Advanced Microbial Bioinformatics Analysis
Why Bioinformatics Matters in Microbial Studies
Sequencing generates vast amounts of raw data, but without the right analysis, those data remain unreadable. At CD Genomics, we combine robust pipelines with advanced statistical and computational tools to help researchers uncover the ecological, functional, and evolutionary significance of microbial communities.
Our Key Bioinformatics Modules
- Diversity Analysis: Alpha and Beta diversity indices, PCA, PCoA, NMDS, and PERMANOVA for statistical comparisons.
- Community Structure Visualization: Intuitive graphics including bar charts, pie charts, heatmaps, Venn diagrams, and Circos plots.
- Environmental Factor Correlation: RDA/CCA, Mantel tests, and MaAsLin for linking microbial shifts to environmental drivers.
- Network and Predictive Modeling: Co-occurrence networks, machine learning (random forest), and ROC analysis for pattern discovery.
- Functional Prediction: Tools such as PICRUSt2, Tax4Fun, BugBase, and FAPROTAX for metabolic and ecological role inference.
- Phylogenetic Analysis: Customizable phylogenetic trees for evolutionary and lineage studies.
 Microbial Diversity Bioinformatics Analysis Overview
Microbial Diversity Bioinformatics Analysis Overview
Takeaway for Researchers:
Sequencing alone reveals who is present, but bioinformatics reveals how communities interact, adapt, and function. By combining advanced algorithms with expert interpretation, CD Genomics ensures that your microbial diversity data become actionable insights for your next publication or research breakthrough.
Summary and Recommendations
Matching Research Goals with the Right Technology
With so many sequencing approaches available, the key is to match your research question with the most suitable method. Below is a quick guide to help you decide:
| Research Purpose | Recommended Solution |
|---|---|
| Quickly understanding microbial structure | 16S/ITS Amplicon Sequencing |
| High-resolution, species-level taxonomy | Full-Length Sequencing or 2bRAD-M |
| Broad ecosystem monitoring (soil, water, air) | eDNA Analysis |
| Absolute microbial load and dynamic shifts | Absolute Quantification Sequencing |
| Functional genes, pathways, resistance profiling | Shotgun Metagenomic Sequencing |
Practical Takeaway
- Use amplicon sequencing for fast, economical snapshots.
- Choose full-length or 2bRAD-M when resolution matters.
- Apply eDNA for non-invasive biodiversity surveys.
- Select absolute quantification to capture real microbial counts.
- Opt for metagenomics when functional or resistance gene insights are required.
Your Partner in Microbial Research
At CD Genomics, we recognize that every project comes with unique challenges—be it sample complexity, budget considerations, or the depth of analysis required. That's why we offer a flexible, multi-dimensional service portfolio backed by expert consultation and advanced bioinformatics support.
If you're unsure which solution best fits your project, our team is here to provide personalized recommendations and experimental design guidance. Contact us today to discuss your microbial diversity research needs.
References
- Huang, X., Zeng, J., Li, S. et al. 16S rRNA, metagenomics and 2bRAD-M sequencing to decode human thanatomicrobiome. Sci Data 11, 736 (2024).
- Díaz, C., Wege, FF., Tang, C.Q. et al. Aquatic suspended particulate matter as source of eDNA for fish metabarcoding. Sci Rep 10, 14352 (2020).
- Sun, Z., Huang, S., Zhu, P., et al. Species-resolved sequencing of low biomass or degraded microbiomes using 2bRAD M. Genome Biology 23, 36 (2022).
- Sethuraman, A., Bowman, B., Eng, K., et al. Analysis of full length 16S genes by single molecule real time sequencing. PacBio (Pacific Biosciences White Paper) (2023).
- Drummond, Alexei J., et al. "Evaluating a multigene environmental DNA approach for biodiversity assessment." GigaScience 4.1 (2015): s13742-015.