Service Overview
Plasmid & BAC Clone Whole-Genome Sequencing is a specialized research service that uses second-generation high-throughput sequencing and third-generation single-molecule sequencing to determine the full-length sequence of plasmids and BAC clones. It enables near-complete or complete circular assemblies by combining Illumina reads with, when needed, long reads and selecting the final sequence based on sequencing depth, GC content, and alignment evidence.
The Challenge of Vector Validation
As plasmids and BAC clones become larger and structurally more complex, simple spot-check or partial sequencing often leaves gaps or ambiguities. Repetitive elements, higher GC regions, and large inserts can all prevent short reads alone from resolving the true construct architecture.
Traditional short-read-only strategies may struggle because:
- Repeats and homology segments can fragment de novo assemblies.
- Size increases (for example, constructs above 20 kb) make closure more difficult.
- Local verification does not guarantee full-length structural integrity.
The CD Genomics Solution
We apply a genome-style approach to plasmid and BAC clone sequencing:
- For suitable smaller constructs, Illumina-only sequencing and assembly can deliver draft maps close to completion, depending on sample complexity.
- For larger or more complex constructs, we recommend a completion strategy similar to bacterial complete-genome workflows, combining Illumina short reads with long reads (PacBio Sequel II or Nanopore).
Final sequence selection is explicitly supported by:
- Sequencing depth
- GC content
- Sequence alignment results
This evidence-based framework helps identify the most plausible full-length plasmid/BAC sequence.
Sequencing Strategies & Technical Specifications
We provide tiered strategies that reflect the size and complexity of your plasmid or BAC clone, based on your internal technical guidance.
How to Choose Illumina-Only vs Hybrid Completion
Use construct size and complexity as the primary guide:
-
For plasmids or BAC clones up to about 20 kb:
High-depth Illumina-only sequencing is usually the first choice for draft maps that may approach completion. -
For constructs larger than about 20 kb or known to be structurally complex or repeat-rich:
Choose a hybrid completion-style package that follows a bacterial complete-genome strategy (Illumina short reads plus long reads such as PacBio Sequel II or Nanopore).
In all cases, the final plasmid/BAC sequence is selected using a combination of sequencing depth, GC content, and sequence alignment results.
Strategy A: High-Depth Illumina (Small Plasmids < 20 kb)
For smaller plasmids and BAC clones within the ≤20 kb range, Illumina NovaSeq sequencing offers a straightforward option for high-depth coverage and draft maps:
- Method: Illumina paired-end sequencing (PE150 or PE250).
- Library insert size: 400–500 bp.
- Depth: Package-defined ~100× coverage.
- Outcome: High-depth draft maps; many smaller constructs can approach completion, but repeat-rich regions may still result in multiple contigs.
Strategy B: Hybrid Assembly (Large Vectors > 20 kb & BACs)
This is the industry gold standard for Complete Maps. For BACs (often 100–300 kb) or plasmids containing extensive homologous regions, we combine platforms to achieve 0 Gaps.
- The "Scaffold": PacBio Sequel II (HiFi reads) or Nanopore reads provide long-range information to span repeats and circularize the sequence.
- The "Polish": Illumina reads are mapped back to the assembly to correct any homopolymer errors inherent in long-read technologies.
- Outcome: A single, circular contig representing the true physical structure of the molecule.
Technical Specification Matrix
| Feature | Bacteria Frame Map A | Bacteria Frame Map B | Complete Map A (Hybrid–PacBio) | Complete Map B (Hybrid–Nanopore) |
|---|---|---|---|---|
| Vector size | Typically ≤ 20 kb | Typically ≤ 20 kb | Often > 20 kb / BAC clones | Often > 20 kb / BAC clones |
| Primary platform | Illumina NovaSeq | Illumina NovaSeq | NovaSeq + PacBio Sequel II | NovaSeq + Nanopore |
| Sequencing mode | PE250 | PE150 | PE150 + long reads | PE150 + long reads |
| Sequencing depth | ~100× | ~100× | ~100× (hybrid) | ~100× (hybrid) |
| Insert size | 400–500 bp | 400–500 bp | ~10 kb (long-read library) | ~10 kb (long-read library) |
| Deliverable goal | Draft map (data volume) | Draft map (data volume) | 0 gaps (completion target) | 0 gaps (completion target) |
“0 gaps” reflects the completion target defined in your internal packages; actual closure depends on sample-specific complexity and data quality.
Key Research Applications
Our Plasmid and BAC sequencing services are utilized by leading academic institutions and biotechnology companies for:
1. Vector Construction Verification
Development of complex vectors often involves large plasmids with inverted terminal repeats (ITRs) or extensive homology regions. Our Hybrid approach resolves these structures, ensuring the research construct is intact and matches the theoretical design before downstream experiments.
2. Metabolic Engineering & Synthetic Biology
When stacking multiple pathways into a single BAC for industrial strain engineering (e.g., metabolite production in Streptomyces or E. coli), structural stability is key. We verify that gene clusters are correctly oriented and free of unintended recombination events.
3. BAC Library Screening (Agriculture)
For plant and animal genomics, BAC libraries are used to map large genomes. We provide high-throughput sequencing of individual BAC clones to assist in physical map construction and positional cloning of trait-specific genes.
4. Microbial Genetics Research
Plasmids are the primary vehicles for Horizontal Gene Transfer (HGT). Our service creates complete maps of plasmids from environmental or laboratory strains, enabling researchers to study the evolution of mobile genetic elements and plasmid incompatibility groups.
Standard Operation Workflow
Our end-to-end pipeline ensures sample integrity and data reliability.
Step 1: Sample Reception & QC
Upon receipt, we perform rigorous Quality Control on your purified DNA using Qubit (concentration) and Agarose Gel Electrophoresis (integrity). We check for degradation and host genomic DNA contamination.
Step 2: Library Construction
- Short Read Library: DNA is fragmented (350bp or 500bp), end-repaired, and adapter-ligated.
- Long Read Library: For PacBio/Nanopore, high-molecular-weight DNA is sheared to ~10-20kb to maximize read length.
Step 3: High-Throughput Sequencing
Samples are sequenced on the respective platforms (NovaSeq/Sequel II/PromethION) to achieve the targeted ~100x depth.
Step 4: Assembly & Gap Filling
We utilize advanced assemblers to assemble reads.
- Gap Filling: Remaining gaps are closed using local assembly or PCR-based verification if necessary.
- Circularization: The software checks for overlapping ends to confirm circularity.
Step 5: Bioinformatic Analysis
Final assembled sequences undergo functional annotation and comparative analysis.

Bioinformatics & Analysis Pipeline
We provide publication-ready figures and comprehensive data files. Our bioinformatics analysis is divided into Standard and Advanced tiers.
Standard Analysis Pipeline
- Data Quality Control: Removal of low-quality reads and adapters.
- Genome Assembly: De novo assembly and scaffolding.
- Coding Gene Prediction: Identification of Open Reading Frames (ORFs) using tools like GeneMark or Prodigal.
- Non-Coding RNA Prediction: Detection of tRNA (tRNAscan-SE) and rRNA (rRNAmmer).
- Structure Prediction: Identification of CRISPRs arrays and Repeat Sequences (Interspersed/Tandem).
Advanced Functional Annotation
- Database Mapping: Annotation against NR (Non-Redundant), GO (Gene Ontology), KEGG (Pathways), eggNOG (Orthologs), and Swiss-Prot.
- Functional Features: Prediction of specific factors tailored to the organism type.
- Protein Features: Signal peptide (SignalP) and Transmembrane helices (TMHMM) prediction.
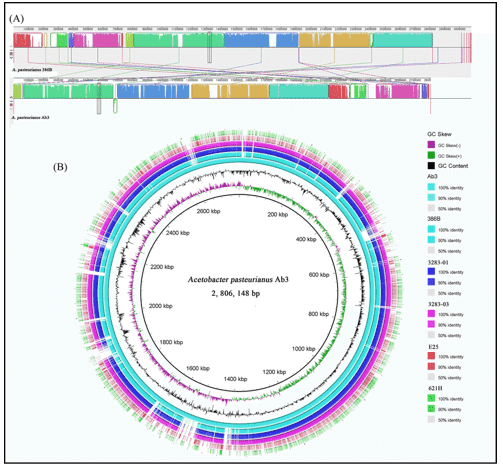
- Genome Visualization: Creation of circular genome maps showing GC skew, gene density, and annotations.
Comparative Genomics (Multi-Sample)
For projects with ≥ 1 reference genome:
- Synteny Analysis: Whole-genome alignment using Mauve or Mummer to visualize rearrangements and homology.
- Pan-Genome Analysis: Defining core and accessory genes across multiple plasmids.
- Phylogenetics: Construction of phylogenetic trees based on single-copy marker genes to trace evolutionary history.
Sample Submission Guidelines
To guarantee "Complete Map" (0 Gap) results, high-quality input material is non-negotiable. Please adhere to the following standards.
| Analysis Type | Minimum Mass | Recommended Mass | Sample Type | Transport |
|---|---|---|---|---|
| Draft Map (Illumina) | 0.2 µg | 0.6 µg | Purified DNA | Dry Ice / Ice Packs |
| Complete Map (PacBio) | 5 µg | 15 µg | Purified DNA | Dry Ice / Ice Packs |
| Complete Map (Nanopore) | 3 µg | 9 µg | Purified DNA | Dry Ice / Ice Packs |
Critical Submission Notes:
- Purity is Paramount: OD260/280 should be between 1.8 and 2.0.
- No Raw Cultures: We do not accept bacterial liquid cultures or plates for this specific service. You must perform the plasmid extraction (Maxi/Midi prep recommended).
- Host Contamination: Minimize host genomic DNA (gDNA). High gDNA levels waste sequencing reads and complicate assembly.
Case Study: Hybrid Assembly of Complex Plasmids
To demonstrate the necessity of Hybrid Sequencing for complex vectors, we highlight a study utilizing the methodology we employ (Unicycler/Hybrid Assembly).
Title: Completing bacterial genome assemblies with multiplex MinION sequencing
Source: Wick RR, et al. Microbial Genomics (2017).
Context: The researchers aimed to resolve the genome of Klebsiella pneumoniae, which contains multiple complex plasmids.
The study compared two approaches:
- Illumina Only: Short-read sequencing (similar to our "Draft Map" service).
- Hybrid Assembly: Illumina short reads + Oxford Nanopore long reads (similar to our "Complete Map B" service).
- Illumina Only: The assembly resulted in a fragmented graph. The plasmid sequences were broken into multiple contigs due to repetitive elements (IS sequences) that the short reads could not bridge.
- Hybrid Approach: The addition of long reads resolved these repeats. The software (Unicycler) successfully produced single, circular contigs for the chromosome and all associated plasmids.

This case confirms that for complete structural verification of plasmids >20kb or those containing repetitive elements, Hybrid Sequencing is the only reliable method to achieve 0 gaps.
Comparison: Why Choose CD Genomics?
| Feature | Traditional Sanger (Primer Walking) | CD Genomics Hybrid WGS |
|---|---|---|
| Throughput | Very Low (1 read/reaction) | Ultra-High (Millions of reads) |
| Assembly Capability | Fails on Repeats & Hairpins | Resolves Repeats (Long Reads) |
| Completeness | Often Fragmented | 0-Gap Circular Map Guaranteed |
| Detection Power | Targeted regions only | Whole Vector (Backbone + Insert) |
| Cost | Expensive for vectors >10kb | Cost-Effective for Large/Multiple Vectors |
| Deliverable | Raw AB1 files + Consensus | Annotated GenBank/Fasta Files |
Frequently Asked Questions (FAQ)
Reference
- Wick RR, Judd LM, Gorrie CL, Holt KE. Completing bacterial genome assemblies with multiplex MinION sequencing. Microbial Genomics. 2017;3(10).











 "
width="400" height="200" loading="lazy"
alt="Bacterial Genome Sequencing for Pollutant Degradation: From Single Strains to Multi-Omics Insights">
"
width="400" height="200" loading="lazy"
alt="Bacterial Genome Sequencing for Pollutant Degradation: From Single Strains to Multi-Omics Insights">