Fungi are incredibly diverse and essential organisms found everywhere—from our bodies to deep-sea environments. They play key roles in nature, such as recycling nutrients and supporting plant growth, and are also widely used in industries like food and medicine. However, many fungi are hard to study because they can't be grown in the lab. This review introduces a powerful genetic method called ITS sequencing (Internal Transcribed Spacer), which has greatly improved how we study fungi. We'll explain what the ITS region is, how the sequencing works, and how new "full-length" technologies are giving us a clearer picture of fungal communities and their impact on the world.
Introduction to Fungal Diversity and Identification Challenges
Fungi make up one of the largest groups of living organisms, with an estimated 1.5 to 3.5 million species. They are vital to ecosystems, especially for breaking down dead matter and cycling nutrients. Fungi also help plants grow through partnerships like mycorrhizae, and they are used in making bread, beer, cheese, antibiotics, and enzymes. However, some fungi can also be harmful—they cause diseases in humans, animals, and plants, and some produce toxins in food.
Despite their importance, fungi have long been difficult to study. Traditional methods—like growing fungi in a lab or examining their shape under a microscope—have major limitations. Many fungi can't be cultured, and their appearance can vary widely, making identification tricky and slow. To overcome these issues, scientists now use DNA-based methods. One of the most effective tools for identifying fungi is analyzing a DNA segment called the Internal Transcribed Spacer (ITS).
The Internal Transcribed Spacer (ITS): Fungi's Genetic Barcode
Ribosomal RNA Gene Structure and the ITS Position
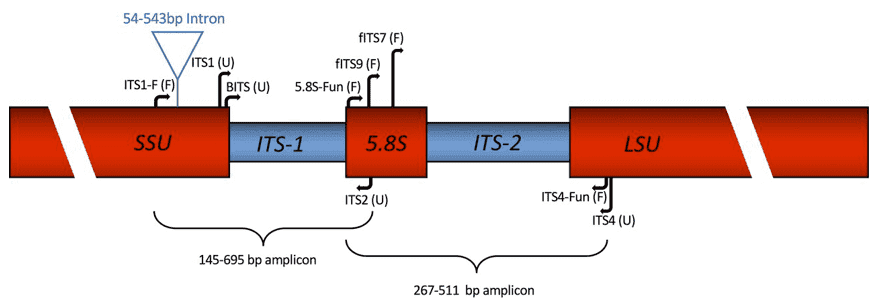
Inside cells, ribosomes are the machines that make proteins. The genetic instructions for making ribosomes come from special DNA regions called rRNA genes, which are repeated many times in fungal genomes. These include three main genes—18S, 5.8S, and 28S—separated by non-coding regions.
The ITS region is one of these non-coding sections, sitting between the 18S and 28S genes. It includes:
- ITS1: between 18S and 5.8S
- ITS2: between 5.8S and 28S
These parts are all copied together into RNA before being processed into mature ribosomal units. The combined ITS1 and ITS2 regions are usually 450–750 base pairs long in fungi.
 Figure 1. Diagram of primer locations in the ribosomal cassette consisting of SSU, ITS1, 5.8S, ITS2, and LSU rDNA.( Martin, K. J. et al, 2005)
Figure 1. Diagram of primer locations in the ribosomal cassette consisting of SSU, ITS1, 5.8S, ITS2, and LSU rDNA.( Martin, K. J. et al, 2005)
Why the ITS Region Is So Useful for Identifying Fungi
The ITS region is a favorite among researchers for several reasons:
- It changes quickly over time: Since ITS doesn't code for proteins, it can accumulate mutations more freely. This makes it great for distinguishing closely related species.
- It's easy to find: Because fungi have many copies of rRNA genes, even tiny DNA samples can be used to amplify the ITS region.
- It balances similarity and difference: ITS is similar enough within a species to group samples together, but different enough between species to tell them apart.
Thanks to these features, in 2012, scientists officially declared the ITS region the "barcode" for fungi. This led to large public databases like GenBank, UNITE, and BOLD that contain countless ITS sequences for researchers to compare and use.
- Strengths:
Great for telling similar species apart
Easy to amplify with standard primers
Supported by large, reliable databases
- Limitations:
In complex samples, some fungal groups may be over- or under-represented due to differences in ITS length or primer bias.
Some public database entries may be misidentified or incomplete.
For difficult groups like Aspergillus, Fusarium, or Penicillium, ITS alone may not be enough. Other DNA markers may be needed for clearer results.
How ITS Works Together with LSU and SSU
While ITS is great for identifying species, it's not ideal for understanding broad evolutionary relationships. For that, scientists use more conserved DNA regions like the large subunit (LSU, or 28S) and small subunit (SSU, or 18S). Combining ITS with these regions gives a fuller picture—from family trees to individual species differences.
Evolution of Sequencing Technologies for Fungal ITS Analysis
1. First-Generation Sequencing (Sanger)
Sanger sequencing was the first widely used method for DNA analysis. It's very accurate but low in throughput. It works well for individual fungal samples but can't handle mixed samples, since it gives overlapping results when multiple species are present.
2. Second-Generation Sequencing (Illumina)
Next-generation sequencing (NGS) technologies like Illumina revolutionized fungal research. These platforms generate millions of short DNA reads quickly and cheaply. Researchers could now study hundreds of fungal samples at once. But there was a catch: the ITS region is longer than these short reads. As a result, scientists often had to choose just part of the ITS region (e.g., ITS1 or ITS2), which limited their ability to distinguish similar species or find new ones.
3. Third-Generation Full-Length ITS Sequencing (PacBio and Oxford Nanopore)
Newer technologies, called third-generation sequencing, combine the accuracy of Sanger with the high output of NGS—and they can read much longer DNA strands.
3.1 PacBio (Pacific Biosciences)
PacBio uses a method where the same DNA strand is read multiple times to ensure accuracy. It can sequence the entire ITS region in one go, removing the need to split the region into parts. This allows for more reliable species identification, even in complex fungal communities.
3.2 Oxford Nanopore Technologies (ONT)
ONT offers ultra-long reads and portable devices. It works by reading DNA as it passes through tiny pores, detecting changes in electrical signals. ONT can read the entire ITS region at once and provides results in real time, which is useful for rapid fieldwork or medical diagnostics.
 Figure 2. Overview of DNA metabarcoding workflow.( Ohta, A. et al, 2023)
Figure 2. Overview of DNA metabarcoding workflow.( Ohta, A. et al, 2023)
3.3 Why Full-Length ITS Sequencing Matters
- Better Species Identification: Capturing the full ITS sequence helps clearly separate closely related species.
- More Accurate Diversity Data: Scientists can measure both species richness (how many species) and evenness (how balanced they are) more reliably.
- Fewer Mistakes in Databases: Full-length sequences offer more unique data, making it easier to match samples correctly and discover unknown species.
Applications of Full-Length ITS Sequencing in Fungal Diversity Research
The power of full-length ITS sequencing has greatly expanded how researchers study fungi across many different fields.
Ecological Surveys of Environmental Fungal Diversity
Full-length ITS sequencing has transformed our ability to explore fungal communities in various environments—such as soil, water, air, and even extreme ecosystems like deserts or deep oceans. This method allows scientists to study how fungi contribute to important processes like nutrient cycling, organic matter breakdown, and symbiosis with plants. It also helps track how environmental changes (like pollution or climate shifts) affect fungal communities, which is valuable for monitoring ecosystem health. By providing a detailed and accurate picture of fungal diversity, this technology has led to the discovery of many previously unknown species.
Rapid Identification of Clinical Pathogenic Fungi
In medicine, quickly and accurately identifying fungal pathogens is critical—especially for patients with weakened immune systems who are vulnerable to serious fungal infections. Full-length ITS sequencing, particularly using real-time platforms like Oxford Nanopore, allows clinicians to identify fungal pathogens directly from patient samples such as blood or tissue, without waiting for cultures to grow. This rapid detection can significantly improve treatment decisions and patient outcomes. It is also useful in detecting unfamiliar or difficult-to-classify fungal species.
 Figure 3. Next-generation Sequencing workflow for fungal diagnosis.( Naik, S. et al, 2024)
Figure 3. Next-generation Sequencing workflow for fungal diagnosis.( Naik, S. et al, 2024)
Fungal Monitoring in Food and Fermentation Industries
Fungi play both helpful and harmful roles in food production. On one hand, they are essential to fermentation processes (like in bread, cheese, or tea); on the other hand, they can cause spoilage or produce toxins harmful to humans. Full-length ITS sequencing makes it possible to precisely track fungal species throughout food production and storage. This helps producers ensure product quality and detect harmful contaminants like Aspergillus, Fusarium, or Penicillium, which can affect both safety and shelf life.
Other Applications
In addition to the above, full-length ITS sequencing is being used in:
- 1) Plant Pathology: To identify fungal diseases in crops and improve agricultural disease management.
- 2) Bioprospecting: To discover new fungi that may produce useful compounds like antibiotics or industrial enzymes.
- 3) Bioremediation: To study fungi that can help break down environmental pollutants.
Challenges and Future Directions
Despite major progress, there are still challenges to overcome and new directions for fungal research using full-length ITS sequencing.
Building Better Databases
Although large databases like UNITE and GenBank hold millions of fungal ITS sequences, they are not perfect. Some entries are mislabeled or lack proper reference data, which can lead to incorrect species identification. More efforts are needed to curate, standardize, and expand these databases, including linking DNA data to actual specimen records and ecological information.
High Diversity, Low Annotation Rates
Even with full-length sequences, many fungi remain unnamed or unclassified because they haven't been studied or described in scientific literature. This means many ITS sequences don't have a matching entry in current databases. Continued efforts in isolating fungi, studying their physical traits, and officially describing new species are crucial for filling these knowledge gaps
Combining with Other "Omics"
To understand not just which fungi are present but also what they're doing, researchers are now combining ITS sequencing with other advanced techniques:
- 1) Metagenomics: To find out what genes are present and what they might do.
- 2) Metatranscriptomics: To see which genes are actively being used in a community.
- 3) Metaproteomics & Metabolomics: To measure actual proteins and chemicals fungi produce.
This "multi-omics" approach helps link fungal identity with real-world biological activity, giving a much fuller picture of their roles in ecosystems.
Quantitative Accuracy and Better Bioinformatics
ITS sequencing mainly shows relative abundance—how common one species is compared to others—but doesn't always give precise counts. Improving methods for absolute quantification remains an active area of research. Additionally, as sequencing produces more data, there's a growing need for user-friendly and accurate bioinformatics tools to handle long-read sequences, reconstruct phylogenies, and make functional predictions.
Conclusion
The Internal Transcribed Spacer (ITS) region is now widely recognized as the genetic "barcode" for fungi, thanks to its ability to distinguish species with high accuracy. With the rise of third-generation sequencing technologies like PacBio and Oxford Nanopore, researchers can now obtain full-length ITS sequences that capture complete genetic variation in one read. This overcomes many of the limitations seen with older techniques.
Full-length ITS sequencing is already making a major impact in ecological surveys, medical diagnostics, food safety, and beyond. Though challenges remain—especially in database quality and the sheer number of unclassified species—the future is promising. By combining full-length ITS data with other molecular tools and "omics" technologies, scientists are poised to gain a much deeper and more functional understanding of the fungal kingdom. These insights will have far-reaching effects across environmental science, agriculture, biotechnology, and human health.
References
- Martin, K. J., & Rygiewicz, P. T. (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC microbiology, 5, 28. https://doi.org/10.1186/1471-2180-5-28
- Ohta, A., Nishi, K., Hirota, K., & Matsuo, Y. (2023). Using nanopore sequencing to identify fungi from clinical samples with high phylogenetic resolution. Scientific reports, 13(1), 9785. https://doi.org/10.1038/s41598-023-37016-0
- Naik, S., Kashyap, D., Deep, J., Darwish, S., Cross, J., Mansoor, E., Garg, V. K., & Honnavar, P. (2024). Utilizing Next-Generation Sequencing: Advancements in the Diagnosis of Fungal Infections. Diagnostics (Basel, Switzerland), 14(15), 1664. https://doi.org/10.3390/diagnostics14151664