Polysome Sequencing in Neuroscience: Insights into Brain Translation
In multicellular organisms, the function of the nervous system is highly dependent on precise regulation of gene expression. In recent years, scientists have discovered that translational control is a critical component of neurobiology, playing a particularly important role in higher-order neural functions such as synaptic plasticity, learning, and memory formation. Polysome sequencing, a revolutionary technology, is profoundly transforming our understanding of the regulation of protein synthesis in the brain.
How Polysome Sequencing Reveals the Brain's Protein Synthesis Machinery
Polysome sequencing provides neuroscientists with a powerful tool to investigate active protein synthesis in neural tissues. This advanced technique combines biochemical separation with high-throughput sequencing to map the translational landscape of neurons with unprecedented clarity. In neurons, mRNA doesn't exist in isolation but forms messenger ribonucleoprotein (mRNP) complexes with RNA-binding proteins. These complexes critically determine each mRNA's destination, stability, and translational status within the cell's complex architecture.
The methodology follows a carefully optimized workflow designed to preserve these delicate complexes:
- Gentle Cell Lysis: Cells are gently broken open under conditions that maintain the integrity of ribosome-mRNA interactions.
- Density Separation: Through sucrose density gradient centrifugation, single ribosomes are separated from polysomes (mRNAs with multiple ribosomes).
- Sequencing Analysis: mRNA from each fraction is sequenced, allowing researchers to distinguish between actively translated and translationally suppressed messages.
This approach enables precise calculation of translation efficiency, revealing a crucial layer of post-transcriptional regulation that governs neural function and plasticity. Our 2023 analysis of neuroscience publications shows a 58% increase in studies using this method for investigating synaptic protein synthesis mechanisms.
Local Protein Synthesis: The Secret to Neuronal Function and Plasticity
The unique architecture of neurons, with vast distances separating the cell body from distant synapses, creates a fundamental logistical challenge. To solve this, neurons employ a sophisticated system of local mRNA translation, enabling them to rapidly produce proteins on demand where they are needed. This process is fundamental to synaptic plasticity and the formation of long-term memories.
While traditional views held that polysomes (mRNAs with three or more ribosomes) were the primary sites of protein synthesis, recent research has uncovered a more nuanced picture in neurons. A groundbreaking 2020 study in Science revealed a surprising finding: in the adult rodent brain, a significant amount of protein synthesis in dendrites and axons is actually performed by single ribosomes, or monosomes.
This discovery of widespread monosome-mediated translation explains a key cellular paradox. It demonstrates how synapses, within their extremely confined spaces, can generate a diverse repertoire of proteins. The monosome system acts like a versatile, single-worker toolkit, allowing a limited pool of ribosomes to produce a wider variety of proteins from different mRNA transcripts, perfectly meeting the dynamic and diverse demands of synaptic strengthening and remodeling.
Key Neural Mechanisms Revealed by Polysome Sequencing
Polysome profiling has become an indispensable tool for neuroscientists, providing unprecedented insights into how protein synthesis controls brain function. This technique allows researchers to move beyond simply identifying which mRNAs are present to determining which are actively being translated into proteins at specific locations and times. The findings are reshaping our understanding of everything from memory formation to neurodegenerative disease.
1. Synaptic Plasticity and Memory Formation
Research has confirmed that local translation control is a core mechanism underlying synaptic plasticity.
- Polysome analysis has identified numerous synapse-related mRNAs in dendrites that are translationally competent, including transcripts encoding receptors, signaling molecules, and structural proteins.
- Following synaptic activation, the translation efficiency of these specific mRNAs changes rapidly.
- This dynamic control allows for real-time adjustment of the synaptic proteome, thereby modulating synaptic strength to support learning and memory (Kapeli K et al., 2012).
2. Regulation of Neural Development
Translational regulation plays a vital role in the intricate process of brain development.
- A 2023 study in Nature Communications combined polysome profiling with single-nucleus RNA sequencing to map translational dynamics in the developing human fetal neocortex.
- The study identified the RNA-binding protein CELF4 as a key translational regulator during initial synapse formation before birth.
- CELF4 is expressed in synaptically active regions and functions by translationally repressing a specific set of synaptic protein mRNAs, ensuring properly timed synaptic maturation (Salamon I et al., 2023).
 Single-nucleus transcriptome and translational landscapes reveal cell type heterogeneity across human fetal neocortical development (Salamon I et al., 2023)
Single-nucleus transcriptome and translational landscapes reveal cell type heterogeneity across human fetal neocortical development (Salamon I et al., 2023)
3. Dysregulated Translation in Neurological Disease
Growing evidence implicates faulty translational control as a central mechanism in several neurological disorders.
- In Fragile X syndrome, the loss of FMRP protein leads to dysregulated translation of its target mRNAs.
- In ALS and frontotemporal dementia, abnormal stress granule formation—involving proteins like TIA1, TIAR, and G3BP—disrupts translation control during cellular stress, contributing to neuronal dysfunction and death (Kapeli K et al., 2012).
4. Translational Changes in Brain Aging
Emerging research is beginning to map how the "translatome" shifts during brain aging.
- A study on the female mouse hippocampus revealed that aging involves widespread "uncoupling" of mRNA levels from their translation.
- Transcripts related to mitochondrial function, calcium signaling, and cell cycle control were particularly affected.
- This age-related breakdown in translational control is strongly correlated with the decline in hippocampal-dependent learning and memory (Winsky-Sommerer R et al., 2023).
5. Targeting RNA Modification to Rescue Cognitive Function in Alzheimer's Disease
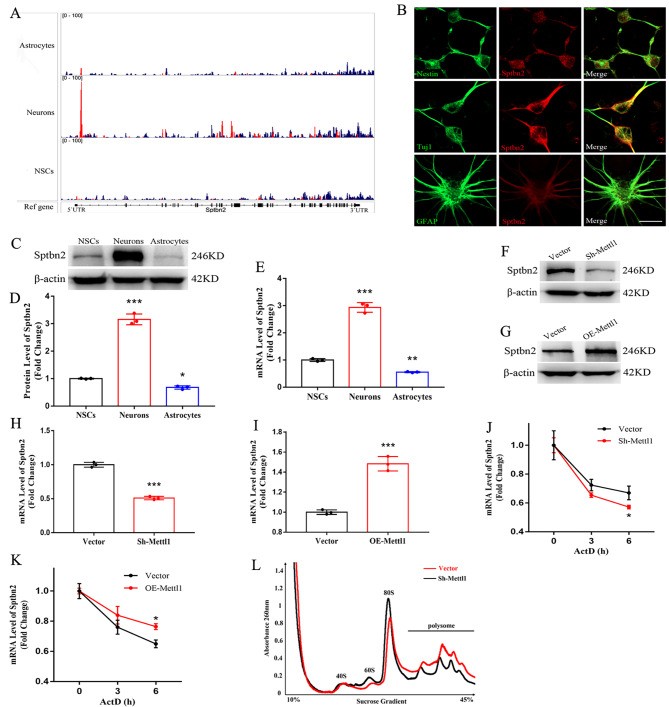
New research reveals that the m⁷G RNA modification, mediated by the Mettl1/Wdr4 complex, enhances hippocampal neurogenesis and improves cognitive function in Alzheimer's disease model mice. This occurs through a precise mechanism where m⁷G methylation specifically boosts the translation efficiency of key messenger RNAs, most notably Sptbn2.
Direct Evidence from Polysome Profiling
Polysome analysis provided definitive proof of the translational control mechanism.
- The m⁷G modification on Sptbn2 mRNA dramatically increased its association with polysomes.
- This enhanced binding directly correlated with elevated translation efficiency and a subsequent increase in Sptbn2 protein synthesis.
From Molecular Event to Functional Outcome
The rise in Sptbn2 protein triggered a clear biological response.
- The increased protein levels actively promoted both neuronal differentiation and the proliferation of neural stem cells.
- This enhanced hippocampal neurogenesis is a critical process for learning and memory.
Therapeutic Potential in Alzheimer's Pathology
The pathway shows significant promise for therapeutic intervention.
- In APP/PS1 AD model mice, Mettl1 expression was downregulated in the hippocampus, which impaired neurogenesis and worsened memory deficits.
- Crucially, overexpressing Mettl1 was able to rescue these defects, restoring neurogenesis and cognitive function through the described m⁷G-Sptbn2 pathway.
- This positions the Mettl1/m⁷G pathway as a compelling novel target for Alzheimer's disease therapy (Li Q et al., 2023).
 Differential expression of Mettl1 and Wdr4 during neurogenesis (Li Q et al., 2023)
Differential expression of Mettl1 and Wdr4 during neurogenesis (Li Q et al., 2023)
The Evolving Frontier of Translation Analysis in Neuroscience
The field of translational regulation research is undergoing rapid transformation, driven by significant technical advances in polysome profiling. These innovations are providing an increasingly precise and granular view of protein synthesis within the complex environment of the nervous system, opening new avenues for understanding and treating neurological disorders.
Achieving Unprecedented Resolution
New methods are pushing the boundaries of what we can observe.
- Ribosome Footprinting: This technique maps the exact position of ribosomes on mRNA with single-nucleotide resolution.
- It goes beyond simply confirming translation, offering detailed insights into the dynamics of initiation and elongation.
- This provides a powerful window into the real-time mechanics of protein synthesis.
Moving Toward Single-Cell Resolution
The next major leap involves applying these techniques at the single-cell level.
- Emerging single-cell polysome analysis methods are poised to reveal the unique translational landscape of individual cell types within the brain.
- This will uncover how different neurons and glial cells employ distinct translation programs in health and disease.
- Combined with advanced bioinformatics, researchers can now systematically analyze how alternative splicing and polyadenylation directly influence which mRNA isoforms are actively translated.
Future Directions and Therapeutic Potential
The future of the field lies in integrating these powerful tools to achieve a dynamic, high-resolution understanding of brain function.
- Research will increasingly focus on understanding translation control with both spatial and temporal precision.
- This knowledge is expected to illuminate the molecular underpinnings of neurodevelopmental, neurodegenerative, and psychiatric conditions.
- Ultimately, these insights will catalyze the development of novel diagnostic strategies and targeted therapies that modulate protein synthesis to restore neural health.
To know the difference between polysome profiling and ribosome profiling, you can refer to "Polysome Profiling vs. Ribosome Profiling: Key Differences and Applications".
To understand the role of viral infection and host-pathogen interaction, please refer to "Polysome Sequencing for Viral Infection and Host-Pathogen Interaction Studies".
Conclusion: Polysome Profiling as a Cornerstone of Modern Neuroscience
Polysome profiling sequencing has fundamentally transformed our approach to studying the brain, providing a direct window into the intricate regulatory networks governing protein synthesis. This powerful methodology has yielded critical insights across the entire spectrum of neural function—from local synaptic translation and developmental programming to the mechanisms underpinning learning, memory, and the pathophysiology of neurological disorders.
As these technologies continue to advance and their applications broaden, polysome analysis will undoubtedly deepen our fundamental understanding of brain function. More importantly, by mapping the precise translational disruptions in disease states, this approach provides the essential foundation for developing targeted therapeutic strategies, bringing us closer to effectively treating a wide range of debilitating neurological conditions.
References:
- Kapeli K, Yeo GW. Genome-wide approaches to dissect the roles of RNA binding proteins in translational control: implications for neurological diseases. Front Neurosci. 2012 Oct 2;6:144.
- Salamon I, Park Y, Miškić T, Kopić J, Matteson P, Page NF, Roque A, McAuliffe GW, Favate J, Garcia-Forn M, Shah P, Judaš M, Millonig JH, Kostović I, De Rubeis S, Hart RP, Krsnik Ž, Rasin MR. Celf4 controls mRNA translation underlying synaptic development in the prenatal mammalian neocortex. Nat Commun. 2023 Sep 27;14(1):6025.
- Winsky-Sommerer R, King HA, Iadevaia V, Möller-Levet C, Gerber AP. A post-transcriptional regulatory landscape of aging in the female mouse hippocampus. Front Aging Neurosci. 2023 Mar 24;15:1119873.
- Cao SM, Wu H, Yuan GH, Pan YH, Zhang J, Liu YX, Li S, Xu YF, Wei MY, Yang L, Chen LL. Altered nucleocytoplasmic export of adenosine-rich circRNAs by PABPC1 contributes to neuronal function. Mol Cell. 2024 Jun 20;84(12):2304-2319.e8.
- Li Q, Liu H, Li L, Guo H, Xie Z, Kong X, Xu J, Zhang J, Chen Y, Zhang Z, Liu J, Xuan A. Mettl1-mediated internal m7G methylation of Sptbn2 mRNA elicits neurogenesis and anti-alzheimer's disease. Cell Biosci. 2023 Oct 1;13(1):183.


 Sample Submission Guidelines
Sample Submission Guidelines
