Applications of Polysome Sequencing in Cancer Research
Polysome profiling is a translationomics research method that uses sucrose density gradient ultracentrifugation to separate mRNAs bound to varying numbers of ribosomes, providing a direct reflection of the translational state within cells.
Combined with high-throughput sequencing, this technique can accurately assess gene translation efficiency and translational regulation mechanisms under various physiological or pathological conditions.
In cancer research, polysome sequencing has been widely used to reveal translational regulatory mechanisms in key biological processes such as tumorigenesis, drug resistance, and immune evasion.
Unlocking Translation Dynamics: A Guide to Polysome Profiling
Polysome profiling provides a direct method for measuring active protein synthesis within cells. This powerful technique for translation regulation studies reveals which messenger RNAs are being actively translated, not just present. The core principle is elegant: the more ribosomes attached to an mRNA strand, the faster it sediments during centrifugation. This allows researchers to separate and analyze mRNAs based on their translational activity.
A Streamlined Workflow from Cells to Data
The experimental process is designed to capture a snapshot of ongoing translation.
- Cell Treatment and Lysis: Cells are first treated with a translation inhibitor like cycloheximide. This "freezes" ribosomes on their mRNA tracks. Cells are then gently lysed, and the cytoplasmic content is collected.
- Separation by Ultracentrifugation: The lysate is layered onto a sucrose density gradient (typically 10%-50%). During ultracentrifugation, components separate by density—free RNA and single ribosomes settle higher, while heavy polysomes sink further.
- Fraction Collection and Analysis: The polysome-containing fractions are collected. RNA is extracted and can be analyzed by qPCR, microarrays, or sequencing to identify which genes are being actively translated.
Key Advantage: Access to Full-Length Regulatory Context
A major strength of this method is its ability to recover full-length, translating mRNAs, including their untranslated regions (UTRs). This is crucial for analyzing mRNA translation mechanisms, as it allows researchers to investigate how regulatory elements in the UTRs control protein production.
Specific Applications in Cancer Mechanism Research
1. Deciphering How Aberrant Translation Initiation Drives Tumor Progression
A review by Wu Q et al. demonstrated that aberrant translation initiation mediated by the eIF4A helicase is a key mechanism of cancer progression in prostate cancer. eIF4A is overexpressed in prostate cancer and is associated with poor prognosis.
Using polysome profiling and Ribo-seq, studies have shown that eIF4A promotes translation initiation of oncogenic transcripts such as the androgen receptor (AR) and HIF1α by deconstructing RNA secondary structures in the 5′ UTR. Its inhibitor, zotatifin, significantly inhibits tumor growth and exhibits synergistic efficacy when combined with AR inhibitors.
2. Unveiling Phase Separation-Mediated Translational Inhibition and Drug Resistance
Meng F et al. discovered that RIOK1 kinase forms stress granules (SGs) through liquid-liquid phase separation in hepatocellular carcinoma (HCC), thereby inhibiting the translation of the tumor suppressor PTEN.
Polysome analysis revealed that RIOK1 overexpression significantly reduced the proportion of PTEN mRNA in polysome fractions, indicating specific inhibition of its translation. This process activates the pentose phosphate pathway, promoting tumor growth and enhancing resistance to tyrosine kinase inhibitors such as sorafenib. Subsequent studies have shown that the drug chidamide can reverse RIOK1-mediated PTEN inhibition by degrading RIOK1, thereby improving chemotherapy sensitivity.
3. Targeting translation initiation: A new therapy for inhibiting oncogenic mRNA translation
Li Q et al., using polysome analysis, discovered that YTHDF1 specifically regulates the translation efficiency of oncogenic target genes through its m6A binding ability. In liver cancer cells, knockdown of YTHDF1 resulted in the relocation of mRNAs for the key autophagy genes ATG2A and ATG14 from actively translating polysome fractions to non-translating fractions, indicating a translational blockade. Conversely, overexpression of wild-type YTHDF1 promoted the enrichment of these two mRNAs in polysomes, enhancing protein synthesis. The key mechanism is that the m6A binding pocket mutant (YTHDF1-MUT) completely loses this function, proving that YTHDF1 "recruits" ribosomes to target mRNA in a strictly m6A-dependent manner, thereby driving liver cancer progression.
 MeRIP-seq and proteomics identified potential targets of YTHDF1 in HCC (Li Q et al., 2021)
MeRIP-seq and proteomics identified potential targets of YTHDF1 in HCC (Li Q et al., 2021)
4. PUS7-mediated pseudouridylation inhibits gastric cancer progression by activating ALKBH3 translation
Chang Y et al. found that the expression of the pseudouridine synthase PUS7 was significantly downregulated in gastric cancer tissues. Functional experiments demonstrated that PUS7 inhibits tumor growth through its catalytic activity. Mechanistically, PUS7 pseudouridylates ALKBH3 mRNA at position U696. Polysome analysis confirmed that this modification significantly enhances ALKBH3 mRNA translation efficiency rather than affecting its stability, thereby increasing expression of the ALKBH3 tumor suppressor protein. This study reveals a novel tumor suppression pathway mediated by mRNA pseudouridylation and provides a potential target for gastric cancer treatment.
5. Exploring the Translational Function of Noncoding RNAs
Polysome analysis can be used to identify the translational potential of noncoding RNAs, such as circRNAs and lncRNAs. For example, Jiang X et al. found that the circular RNA hsa_circ_0000467 is significantly overexpressed in colorectal cancer and associated with poor prognosis. Functionally, it promotes cancer cell growth and metastasis. Polysome analysis revealed its core mechanism: hsa_circ_0000467 acts as a molecular scaffold in the cytoplasm, binding to the RNA helicase eIF4A3 and c-Myc mRNA to form a ternary complex, significantly enhancing c-Myc translation efficiency without affecting its mRNA level. Elevated c-Myc protein, in turn, regulates downstream cell cycle and EMT-related targets, ultimately driving tumor progression. This study provides new potential diagnostic markers and therapeutic targets for colorectal cancer.
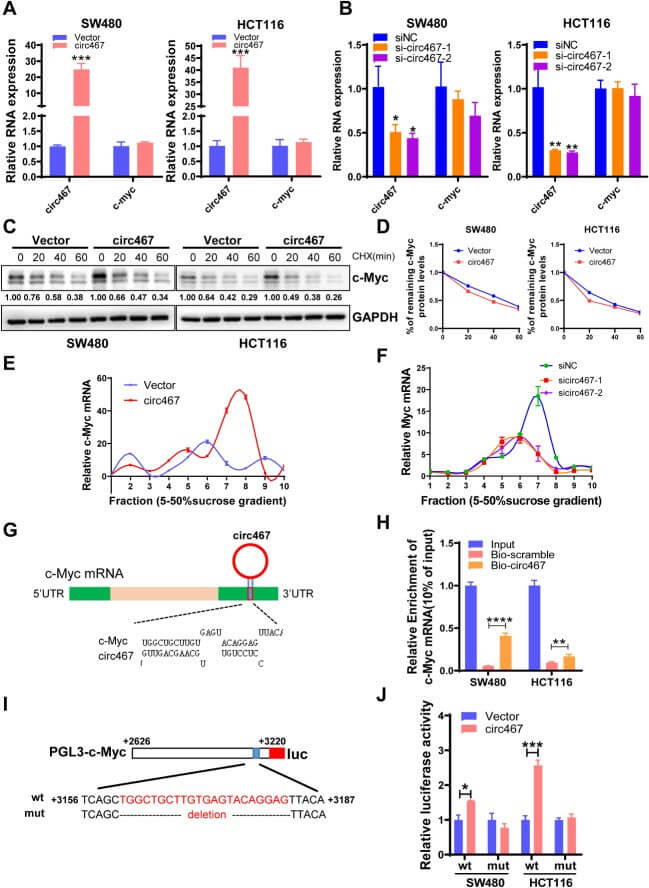 Circ467 may increase the translation efficiency of c-Myc (Jiang X et al., 2024)
Circ467 may increase the translation efficiency of c-Myc (Jiang X et al., 2024)
6. Driving Glucose Metabolism Reprogramming to Promote Liver Cancer Progression
Xia P et al. discovered that the methyltransferase METTL5 is highly expressed in liver cancer and can drive glucose metabolism reprogramming (Warburg effect). Mechanistically, polysome analysis revealed that METTL5 significantly enhances the mRNA translation of the deubiquitinating enzyme USP5, thereby increasing USP5 protein levels. USP5 inhibits the K48-linked polyubiquitination and degradation of c-Myc, enhancing its stability and thereby activating the expression of glycolytic genes such as LDHA and PKM2, ultimately promoting liver cancer proliferation and metastasis. Studies have confirmed that CREB1/P300 is an upstream factor regulating METTL5 transcription. Knockdown of METTL5 in PDX models demonstrated a significant antitumor effect, suggesting its potential as a new therapeutic target for liver cancer.
7. LIN28B drives liver cancer initiation through a translational cascade regulating an RBP network
Hsieh MH et al. reveals that LIN28B is a key driver of liver cancer initiation. Its core mechanism is not primarily through inhibition of let-7, but rather through direct enhancement of mRNA translation of eight key members of a core regulatory network of 15 RNA-binding proteins (RBPs). Polysome analysis confirmed the direct translational upregulation of this RBP network by LIN28B. This network forms a powerful post-transcriptional regulatory cascade, ultimately promoting processes such as protein synthesis, providing the essential foundation for liver cancer initiation.
To learn about the advanced applications of polysome sequencing in plant research, please refer to "Advanced Applications of Polysome Sequencing in Plant Research".
To understand how polysome sequencing plays a role in viral infection and host-pathogen interactions, refer to "Polysome Sequencing for Viral Infection and Host-Pathogen Interaction Studies".
Technical Comparison and Advantages
| Feature | Polysome Profiling/Seq | Ribo-seq | RNC-seq |
|---|---|---|---|
| Detection Scope | Full-length mRNA (including UTRs) | Ribosome-protected fragments | Full-length mRNA |
| Core Advantages | Directly reflects translational load and efficiency | Precisely locates ribosome positions | Suitable for low-abundance samples |
| Applications in Cancer Research | Assess global changes in translational activity; Analyze translation efficiency of specific oncogenic mRNAs | Discover non-canonical ORFs and frameshift events | Analysis of scarce clinical samples (e.g., biopsy tissues) |
The unique advantage of polysome profiling lies in its ability to directly quantify the number of ribosomes on each mRNA, thereby providing an intuitive reflection of dynamic changes in translation efficiency.
To learn more about the differences and applications of polysome profiling and ribosome profiling, please refer to "Polysome Profiling vs. Ribosome Profiling: Key Differences and Applications".
Services you may be interested in
Translation Control in Cancer: Current Insights and Future Directions
Polysome sequencing is revolutionizing our understanding of cancer by revealing widespread dysregulation at the translation level. This technology provides critical insights into tumor progression, drug resistance, and immune response mechanisms that are often invisible to other omics approaches. For drug developers, analyzing translational control offers a direct window into the functional proteome, uncovering novel therapeutic vulnerabilities.
Looking ahead, the integration of polysome profiling with cutting-edge single-cell and spatial multi-omics platforms will unlock unprecedented precision. A 2023 industry forecast suggests that the market for translation-focused oncology diagnostics will grow by 35% annually, driven by these technological synergies.
Future applications are poised to impact three key areas:
- Clinical Diagnostics: Translatione analysis of minimal clinical samples, like biopsies, could refine cancer subtyping and improve prognostic accuracy.
- Novel Drug Discovery: This approach enables the rational development of therapies targeting translation machinery, such as initiation factors (e.g., eIF4A) or context-specific kinases (e.g., RIOK1).
- Next-Generation Immunotherapies: Understanding how translation errors (e.g., frameshifting) generate neoantigens can inform strategies to boost the efficacy of immune checkpoint inhibitors.
References:
- Wu Q, Tavazoie SF. Ribosomal rebellion: When protein factories drive cancer progression. Cancer Cell. 2025 May 12;43(5):808-809.
- Meng F, Li H, Huang Y, Wang C, Liu Y, Zhang C, Chen D, Zeng T, Zhang S, Li Y, Zhang B, Lang C, Xia J, Xiong W, Pan S, Sun X, Thorne RF, Liu Y, Wang J, Zhang S, Song R, Wang J, Liu L. RIOK1 phase separation restricts PTEN translation via stress granules activating tumor growth in hepatocellular carcinoma. Nat Cancer. 2025 Jul;6(7):1223-1241.
- Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, Hu Y, Qiu J, Pu L, Tang J, Wang X. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021 Feb 23;6(1):76.
- Chang Y, Jin H, Cui Y, Yang F, Chen K, Kuang W, Huo C, Xu Z, Li Y, Lin A, Yang B, Liu W, Xie S, Zhou T. PUS7-dependent pseudouridylation of ALKBH3 mRNA inhibits gastric cancer progression. Clin Transl Med. 2024 Aug;14(8):e1811.
- Xia P, Zhang H, Lu H, Xu K, Jiang X, Jiang Y, Gongye X, Chen Z, Liu J, Chen X, Ma W, Zhang Z, Yuan Y. METTL5 stabilizes c-Myc by facilitating USP5 translation to reprogram glucose metabolism and promote hepatocellular carcinoma progression. Cancer Commun (Lond). 2023 Mar;43(3):338-364.
- Jiang X, Peng M, Liu Q, Peng Q, Oyang L, Li S, Xu X, Shen M, Wang J, Li H, Wu N, Tan S, Lin J, Xia L, Tang Y, Luo X, Liao Q, Zhou Y. Circular RNA hsa_circ_0000467 promotes colorectal cancer progression by promoting eIF4A3-mediated c-Myc translation. Mol Cancer. 2024 Jul 31;23(1):151.
- Hsieh MH, Wei Y, Li L, Nguyen LH, Lin YH, Yong JM, Sun X, Wang X, Luo X, Knutson SK, Bracken C, Daley GQ, Powers JT, Zhu H. Liver cancer initiation requires translational activation by an oncofetal regulon involving LIN28 proteins. J Clin Invest. 2024 Jun 13;134(15):e165734.


 Sample Submission Guidelines
Sample Submission Guidelines
