Polysome Sequencing in Immunology: Translational Regulation of Immune Responses
For decades, immunology research has centered on how genes are transcribed. We now understand that controlling protein synthesis—known as translational regulation—is equally vital for a robust immune response. This is where Polysome sequencing delivers crucial insights.
Polysome sequencing captures a real-time snapshot of protein production by identifying mRNAs actively being translated by ribosomes. This "translational landscape" reveals how immune cells rapidly reprogram their machinery to confront threats.
Why This Matters in Immunology
When faced with an infection, immune cells must act immediately. They cannot wait for new gene transcription. Translational control allows for this instant response.
When T-cells encounter an antigen, they rapidly initiate proliferation and cytokine production. Polysome profiling reveals that this burst of protein synthesis is largely driven by the swift activation of pre-existing mRNA pools—enabling a rapid immune response that circumvents the slower transcription-based gene expression pathway.
By separating and sequencing these active mRNA-ribosome complexes, researchers obtain genome-wide data on translation efficiency. This reveals the sophisticated mechanisms immune cells use to control protein production at this final, decisive stage.
Table: Key Technical Features of Polysome Analysis Technology in Immunology Research
| Technical Feature | Significance in Immunology Research | Application Example |
|---|---|---|
| Provides quantitative translation efficiency data | Reveals discrepancies between transcript expression and protein abundance | Identifying key translationally regulated genes in immune cell differentiation |
| High temporal resolution | Captures dynamic changes in protein synthesis during immune activation | Analyzing translation of immediate-early response genes after T cell stimulation |
| Genome-wide coverage | Systematically discovers translationally regulated networks involved in immune responses | Mapping translational reprogramming within cytokine signaling pathways |
| Multi-omics data from a single experiment | Enables integrated analysis of correlations and differences between transcriptome and translatome | Deciphering the molecular mechanisms of immune checkpoint inhibitor action |
Services you may be interested in
Decoding Immune Cell Development Through Translation Control
Immune cell differentiation depends critically on protein synthesis regulation—the process through which cells prioritize specific proteins from available mRNA templates. This post-transcriptional control mechanism directly shapes stem cell fate decisions, determining whether they preserve their multipotent state or differentiate into specialized immune lineages. Our understanding of this mechanism comes from Polysome sequencing, which captures actively translated mRNAs within living cells.
Research Methodology: From Cells to Insights
The approach begins by isolating specific immune cell populations, including hematopoietic stem cells and myeloid progenitors. Researchers separate active translation complexes and sequence their mRNA content. Bioinformatics analysis then calculates translation efficiency and identifies regulated pathways, revealing how protein synthesis shapes cellular identity.
Key Discovery in Stem Cell Maintenance
Hematopoietic stem cells employ a refined protein synthesis strategy. They maintain generally low translational activity while selectively and efficiently producing proteins critical for self-renewal. This precise gene expression regulation sustains their multipotent capacity through three key mechanisms:
- Oxygen-sensing pathways
- Extracellular signaling systems
- Energy metabolism adaptation
This targeted translation control preserves stem cell identity while preparing for potential differentiation cues.
The Efficiency Advantage in Cellular Regulation
A key discovery reveals how translation efficiency shapes cellular function. Numerous moderately abundant mRNAs achieve substantial protein synthesis through highly efficient translation. These particular transcripts encode essential regulators governing:
- Stem cell maintenance potential
- Reactive oxygen species defense
- Genomic integrity maintenance
- Metabolic and signaling networks
This sophisticated regulatory strategy enables rapid protein production without requiring abundant mRNA templates. This represents a refined control mechanism in cellular development and differentiation. (Spevak CC et al., 2019).
Rapid Protein Synthesis Reprogramming in Lymphocyte Activation
When lymphocytes recognize antigens, their protein synthesis machinery undergoes a fundamental shift. This reprogramming allows T and B cells to quickly proliferate and perform specialized immune functions through coordinated activation of existing mRNA templates—a vital layer of biological control that works alongside DNA-level regulation to coordinate effective immune responses.
Key Signaling Pathways in Translation Control
The rapid expansion of immune cells is coordinated by two complementary regulatory systems. While mTOR signaling generally promotes translation initiation, CDK1 provides an alternative activation pathway when mTOR activity is limited. This regulatory network functions through 4E-BP1 phosphorylation, which releases initiation factors and enables the protein synthesis necessary for immune cell differentiation.
In myeloid precursors, researchers documented a notable adaptation: even with reduced mTOR signaling due to protein degradation, CDK1 successfully sustained protein production through 4E-BP1 modification. This demonstrates how cells maintain essential functions through redundant control mechanisms.
Cell-Type Specific Translation Patterns
Recent polysome profiling studies reveal sophisticated translation control mechanisms in specialized immune cells. The research shows regulatory T cells achieve high FoxP3 protein synthesis through optimized translation efficiency rather than mRNA abundance. Meanwhile, developing B cells significantly increase immunoglobulin production through specialized translation pathways.
These findings highlight how immune cells strategically manage protein synthesis to perform specialized functions. The insights suggest new therapeutic approaches could emerge from targeting these translation control mechanisms. This represents a promising frontier for developing immune-modulating treatments (Spevak CC et al., 2019).
Translational Control in Innate Immunity: Beyond Genetic Regulation
Innate immune cells utilize precise translational control to generate immediate responses against pathogens. This protein synthesis regulation enables frontline defenders to rapidly adjust their functional output without relying on slower genetic adaptations. For immunotherapy developers, these mechanisms reveal new therapeutic potential for precisely modulating immune activity through post-transcriptional pathways.
Professional Antigen-Presenting Cells
Upon pathogen detection, dendritic cells rapidly reshape their protein synthesis landscape to synchronize inflammatory signaling and antigen processing. Concurrently, macrophages develop specialized translation patterns during their differentiation into pro-inflammatory M1 or regenerative M2 states, with emerging evidence pointing to inflammasome-mediated regulation of these translational programs.
Natural Killer Cell Adaptation
Natural killer cells exhibit remarkable adaptability in protein synthesis when exposed to cytokine signals. Specific interleukins trigger distinct responses: IL-2, IL-12 and IL-15 enhance production of cytotoxic proteins, while IL-18 establishes a unique state of multifunctional readiness. During this transition, more than a thousand genes undergo reprogramming at the translation level, affecting cellular proliferation, differentiation and immune regulatory functions.
This cytokine-directed control allows NK cells to precisely calibrate their responses to environmental signals. The findings underscore how protein synthesis regulation provides a crucial strategic dimension to immune control, operating alongside established genetic mechanisms to fine-tune cellular responses (Cui A et al., 2023).
Viral Hijacking of Host Translation: A Molecular Battlefield
The ongoing molecular competition between viruses and host immune defenses reveals sophisticated strategies for controlling cellular protein synthesis. Respiratory syncytial virus (RSV) demonstrates particular finesse in redirecting host translation machinery toward viral protein production. Rather than inducing broad translational arrest as many viruses do, RSV employs a targeted approach that selectively maintains specific host functions while exploiting the protein synthesis apparatus. This nuanced strategy presents both challenges and opportunities for developing targeted antiviral interventions and advancing infectious disease understanding.
RSV's Translation Optimization Strategy
RSV achieves translational dominance through several clever adaptations:
- Viral mRNAs mimic host structures but escape translation inhibition
- Infection triggers redistribution from 80S monosomes to polysomes
- Ribosome loading increases significantly on target mRNAs
- AU-rich transcripts receive preferential ribosomal recruitment
The viral M2-1 protein appears central to this process. Found associated with polysomes, M2-1 likely acts as a delivery factor for viral and host AU-rich transcripts to the translation machinery. This elegant adaptation allows RSV to prioritize its protein synthesis without completely shutting down host translation (Kerkhofs K et al., 2024).
Host Countermeasures: The VISA/MAVS Regulatory System
The immune system has developed sophisticated counter-strategies against viral manipulation of cellular machinery. A key example involves the precise post-translational regulation of the adapter protein VISA/MAVS:
- Tankyrase enzymes (TNKS1/2) modify VISA through PARylation
- E3 ubiquitin ligase RNF146 recognizes these PAR modifications
- K48-linked polyubiquitination marks VISA for proteasomal degradation
- This mechanism provides crucial negative regulation during RNA viral infection
This recently elucidated pathway represents the host's evolutionary adaptation to viral interference, demonstrating the continuous molecular innovation that characterizes host-pathogen interactions (Xu YR et al., 2022).
 Polysome association with mRNA in immune cells during viral infection (Kerkhofs K et al., 2024)
Polysome association with mRNA in immune cells during viral infection (Kerkhofs K et al., 2024)
Immune Cells' Strategic Translation Reprogramming During Viral Infection
When detecting viral invasion, immune cells strategically reprogram their protein synthesis to mount effective defenses. This sophisticated adaptation represents a critical evolutionary development in antiviral immunity. Dendritic cells exemplify this capability by rapidly reshaping their translation profile upon recognizing viral RNA through sensors like protein kinase R (PKR), enabling precise control of immune responses without relying solely on transcriptional changes.
Balanced Translation Control
While PKR activation generally suppresses global protein synthesis through eIF2α phosphorylation, immune cells maintain precise translation of essential defense proteins. This selective mechanism preserves the production of critical immune mediators including type I interferons, key inflammatory cytokines, and other antiviral effectors necessary for an effective host response. The process involves specialized RNA elements and translation initiation mechanisms that bypass conventional regulatory checkpoints, ensuring continuous synthesis of protective proteins despite viral countermeasures.
Rapid Innate Immune Adaptation
Within hours of viral detection, macrophages and dendritic cells undergo significant translation reprogramming. They prioritize inflammatory gene translation while suppressing most housekeeping genes. Polysome sequencing reveals this strategic shift involves not just transcriptional changes but precise control of translation efficiency. This enables rapid establishment of antiviral states while conserving cellular resources for sustained immune challenges (Cui A et al., 2024).
Uncovering Translation Control in Dendritic Cell Immune Activation
Our investigation systematically examined translational regulation during dendritic cell maturation using Polysome Sequencing combined with DNA microarray technology. We tracked how protein synthesis patterns change when immune cells become activated.
Research Approach
We employed a structured methodology to capture dynamic translation changes:
- Derived dendritic cells from human monocytes and stimulated them with LPS
- Collected samples at multiple timepoints (0, 4, and 16 hours) to track progression
- Isolated actively translated mRNAs using sucrose density gradient centrifugation
- Compared total mRNA versus polysome-bound fractions using microarray analysis
Key Discoveries
The study revealed extensive translational control during immune activation:
- Widespread Translation Regulation: We identified 375 transcripts undergoing significant translational changes during DC maturation. This demonstrates that immune response precision extends far beyond transcriptional control to include dynamic translation efficiency adjustments.
- Self-Regulating Protein Synthesis Machinery
- The most significantly regulated pathway involved protein biosynthesis itself. A cluster of 11 large ribosomal protein mRNAs showed particularly interesting behavior:
- Early activation: actively translated with polysome association
- Late maturation (16 hours): significant translation suppression and polysome dissociation
- The most significantly regulated pathway involved protein biosynthesis itself. A cluster of 11 large ribosomal protein mRNAs showed particularly interesting behavior:
- Novel Negative Feedback Circuit: This pattern reveals an elegant feedback mechanism. Cells initially boost translation capacity to meet protein demand during early immune response. Later, they deliberately limit further production by downregulating ribosomal protein synthesis, preventing excessive immune activation and maintaining homeostasis (Ceppi M et al., 2009).
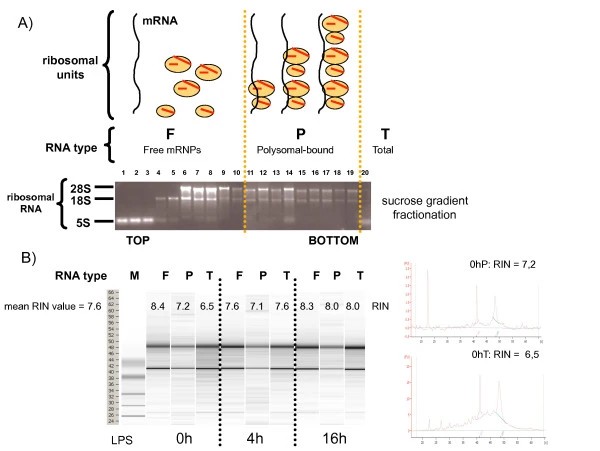 Efficient polysome profiling of human dendritic cells for translational regulation research (Ceppi M et al., 2009)
Efficient polysome profiling of human dendritic cells for translational regulation research (Ceppi M et al., 2009)
Designing Robust Polysome Sequencing Studies for Immune Cell Translation Analysis
Generating reliable translatome data from immune cells requires meticulous experimental planning and execution. Polysome Sequencing offers powerful insights into translational regulation, but its success depends on carefully controlled conditions throughout the workflow.
Experimental Workflow Overview
The standard polysome sequencing approach involves several critical steps:
- Treat cells with cycloheximide to preserve ribosome positions on mRNAs
- Separate polysome fractions using sucrose density gradient centrifugation
- Extract RNA from different polysome populations for sequencing
- Analyze sequencing data to identify actively translated transcripts
Data Analysis Strategy
Translation efficiency is typically calculated as the ratio of polysome-associated mRNA to total mRNA. Comparing these efficiency values across different immune states reveals differentially translated transcripts. Integrated analysis with transcriptomic and proteomic data often shows that translation efficiency changes frequently correlate better with protein expression than mRNA abundance alone.
Key Technical Considerations for Success
Several factors significantly impact Polysome Sequencing data quality in immunology research:
- Cell Population Purity: Studies on rare immune subsets may require fluorescence-activated cell sorting to obtain homogeneous populations
- Temporal Resolution: Immune responses evolve rapidly, so timepoint selection critically influences findings. Early timepoints (e.g., 4 hours post-stimulation) capture immediate translation changes, while later intervals reveal secondary responses
- Activation State Control: Immune cells can activate spontaneously in culture, necessitating proper controls and tightly regulated conditions
- Cell-Type Specific Interpretation: Different immune cells exhibit varying ribosomal densities and translation characteristics, requiring careful data interpretation
These considerations help ensure that observed translation changes genuinely reflect biological regulation rather than technical artifacts.
For more information on the applications of polyribosomal sequencing in neuroscience, please refer to "Polysome Sequencing in Neuroscience: Insights into Brain Translation".
For more information on the applications of polyribosomal sequencing in virus-host interaction studies, please refer to "Polysome Sequencing for Viral Infection and Host-Pathogen Interaction Studies".
To learn about the challenges and limitations of polyribosomal sequencing, please refer to "Challenges and Limitations of Polysome Sequencing".
The Future of Translation Control in Immunology Research and Therapy
Polysome Sequencing continues to reveal unprecedented insights into how immune cells control protein production. As this technology evolves, it opens new possibilities for understanding immune regulation and developing innovative therapies. The integration of translatome data with transcriptomic, proteomic, and epigenetic information will provide increasingly comprehensive views of immune system function.
Emerging Technological Frontiers
Several exciting developments are shaping the future of translational control research:
- Single-cell polysome sequencing will resolve cellular heterogeneity in immune responses
- Multi-omics integration will reveal connections between different regulatory layers
- Advanced computational tools will model complex translational networks
- Dynamic tracking of translation changes will capture immune cell plasticity
Therapeutic Applications and Opportunities
Understanding translational control mechanisms creates new opportunities for clinical intervention:
- mTOR pathway inhibitors could modulate abnormal immune activation
- Translation initiation factors represent potential drug targets
- Fine-tuning translation programs may improve cancer immunotherapy
- Autoimmune conditions might benefit from selective translation modulation
These approaches could enhance treatment efficacy while reducing side effects through more precise immune manipulation.
Concluding Perspective
Polysome analysis has fundamentally expanded our understanding of immune regulation. We now appreciate translation control as equally important as transcriptional regulation in shaping immune responses. As technology advances and datasets grow, we move closer to comprehensive mapping of immune translation networks. This knowledge will undoubtedly inspire new diagnostic and therapeutic strategies for immune-related disorders.
People Also Ask
What is analysis of translation using Polysome sequencing?
Polysome sequencing has been developed to infer the translational status of a specific mRNA species or to analyze the translatome, i.e. the subset of mRNAs actively translated in a cell. Polysome sequencing is especially suitable for emergent model organisms for which genomic data are limited.
What is the principle of Polysome sequencing?
The fundamental principle is to compare the abundance of each mRNA in the heavy, actively translating polysome fractions against its presence in the single ribosome (80S) fraction. This comparison allows you to calculate a translation efficiency (TE) score for each gene.
References:
- Spevak CC, Elias HK, Kannan L, Ali MAE, Martin GH, Selvaraj S, Eng WS, Ernlund A, Rajasekhar VK, Woolthuis CM, Zhao G, Ha CJ, Schneider RJ, Park CY. Hematopoietic Stem and Progenitor Cells Exhibit Stage-Specific Translational Programs via mTOR- and CDK1-Dependent Mechanisms. Cell Stem Cell. 2020 May 7;26(5):755-765.e7.
- Cui A, Huang T, Li S, Ma A, Pérez JL, Sander C, Keskin DB, Wu CJ, Fraenkel E, Hacohen N. Dictionary of immune responses to cytokines at single-cell resolution. Nature. 2024 Jan;625(7994):377-384.
- Kerkhofs K, Guydosh NR, Bayfield MA. Respiratory Syncytial Virus (RSV) optimizes the translational landscape during infection. bioRxiv [Preprint]. 2024 Aug 3:2024.08.02.606199.
- Xu YR, Shi ML, Zhang Y, Kong N, Wang C, Xiao YF, Du SS, Zhu QY, Lei CQ. Tankyrases inhibit innate antiviral response by PARylating VISA/MAVS and priming it for RNF146-mediated ubiquitination and degradation. Proc Natl Acad Sci U S A. 2022 Jun 28;119(26):e2122805119.
- Ceppi M, Clavarino G, Gatti E, Schmidt EK, de Gassart A, Blankenship D, Ogola G, Banchereau J, Chaussabel D, Pierre P. Ribosomal protein mRNAs are translationally-regulated during human dendritic cells activation by LPS. Immunome Res. 2009 Nov 27;5:5.


 Sample Submission Guidelines
Sample Submission Guidelines
