Challenges and Limitations of Polysome Sequencing
Polysome profiling/sequencing, a key technology in translatomics research, can reveal the fine-grained regulation of genes at the protein translation level by analyzing mRNA bound to different numbers of ribosomes. Although it is considered the "gold standard" for assessing translation efficiency, its limitations in practical applications cannot be ignored. The following is a detailed analysis of its main challenges and limitations.
Inherent Challenges in Technical Operation and Sample Processing
Polyribosomal sequencing has a high technical threshold, with challenges beginning in sample preparation and separation.
- Complex Fragility: The binding of ribosomes to mRNA is non-covalent, making the conformation of polyribosomal complexes very fragile.
- During cell disruption, mechanical vibrations or changes in the microenvironment can easily lead to complex dissociation, introducing analytical bias.
- Equipment and Operational Requirements: This technique relies on sucrose density gradient ultracentrifugation for separation.
- Preparing the sucrose gradient is both time-consuming and labor-intensive, and its resolution can vary depending on laboratory conditions and operator skill, directly affecting the reproducibility and comparability of results.
- Large Sample Requirement: To obtain sufficient RNA for subsequent sequencing, a large amount of starting material is typically required, such as at least 1 × 107 cells or 100 mg of tissue. This makes studies on rare cell types or small amounts of clinical samples difficult.
 Strategy for investigating the translational efficiency of mRNAs by polysome profiling and microarray hybridization (Krishnan K et al., 2014)
Strategy for investigating the translational efficiency of mRNAs by polysome profiling and microarray hybridization (Krishnan K et al., 2014)
Inherent Limitations of Resolution and Result Interpretation
Even with the successful isolation of polyribosomes, inherent resolution limitations exist in the result interpretation.
- Risk of Misjudging "Active Translation": Traditionally, mRNAs bound to polyribosomes (mRNAs binding to multiple ribosomes) are considered to represent active translation. However, research shows that mRNAs bound to monoribosomes can also be translationally active. The technology itself may not be able to effectively distinguish between mRNAs bound to monoribosomes and those bound to polyribosomes, leading to misinterpretations of the translational status of specific transcripts.
- Co-migration Interference: Sucrose density gradients may not be effective in separating polyribosomes from other large ribonucleoprotein complexes in the cytoplasm. This co-migration phenomenon can lead to incorrect representations of transcript ribosome occupancy.
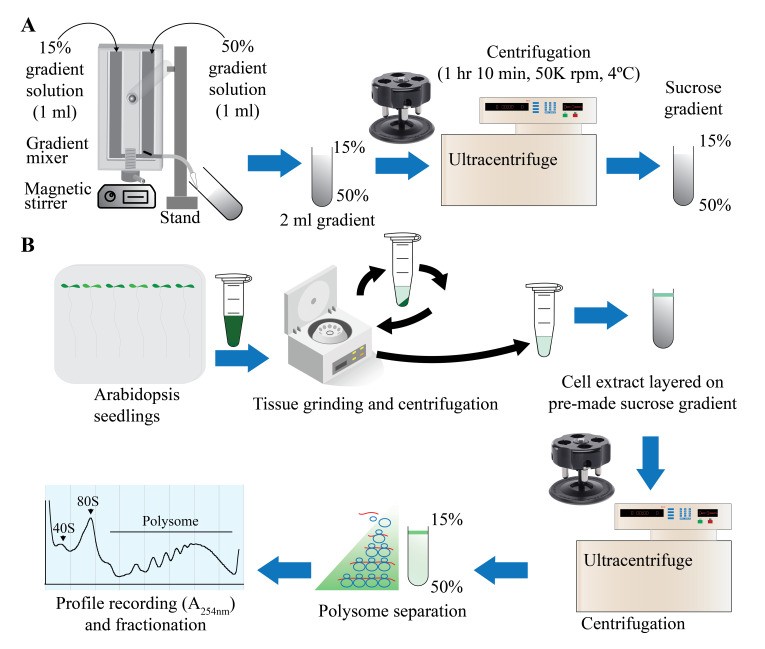 Graphical overview of polysome profiling using mini sucrose gradient (Lokdarshi A et al., 2023)
Graphical overview of polysome profiling using mini sucrose gradient (Lokdarshi A et al., 2023)
The Complexity and Challenges of Data Analysis
After obtaining the raw data, subsequent bioinformatics analysis presents another major challenge.
- It requires a complex bioinformatics workflow: Extracting biologically meaningful information from raw sequencing data requires a series of steps, including data quality control, reference genome alignment, quantitative analysis, differentially translated gene identification, and translation efficiency calculation. For example, calculating translation efficiency (TE) typically requires combining polysome-seq data with ordinary mRNA-seq data.
- Limitations of Short-Read Sequencing: Commonly used short-read sequencing technologies have inherent limitations in accurately identifying alternative splicing sites and resolving complex transcript structures such as circular RNA.
- Misconceptions in Data Interpretation: The sequencing read density of the ribosomal footprint is not directly equivalent to translational activity. It is affected by both the translation initiation rate and the ribosomal elongation rate. For example, if translation stops at a specific mRNA, the ribosomal footprint may be highly enriched, but the actual protein synthesis efficiency may be zero.
For more information on data analysis and interpretation in polyribosome sequencing, please refer to "Data Analysis and Interpretation in Polysome Sequencing".
Application Limitations in Specific Research Scenarioscopy Scenarioscopy
The application of polysome sequencing technology is significantly limited in specific research directions.
- Specific Cell Type Studies: While technologies such as TRAP-seq (translational ribosomal affinity purification sequencing) and RiboTag have been developed for studying the translatome of specific cell types in complex tissues, these methods typically require genetic modification of the organism (e.g., introducing tagged ribosomal proteins), which limits their application in many species that are difficult to modify.
- Background Noise Interference: Even with the above technologies, it is still necessary to minimize background noise from non-target cell transcripts during experiments.
- Limitations for Specific Transcripts: While polysome profiling combined with qPCR or sequencing can be used to identify the translational potential of non-coding RNAs, ribosomal footprinting sequencing may provide higher resolution information for the precise resolution of some non-classical translation events (such as upstream open reading frame translation).
Case Study Analysis: Technical Challenges in Translation Profiling Research
| Case Description | Technical Challenges Involved | Key Findings / Technical Difficulties |
|---|---|---|
| Pancreatic Cancer Drug Resistance Study | Technical Operational Complexity, Data Interpretation | Gemcitabine treatment inhibited global protein synthesis (polysome reduction), but ZEB1 mRNA (shorter 3'UTR isoform) was retained explicitly in polysomes, indicating unique translational regulation. Data analysis required distinguishing this "counter-trend" behavior of specific transcripts. |
| Cardiac Fibrosis Research | Data Analysis Complexity, Translation Efficiency Calculations | To study the EPRS gene's role, joint analysis of Polysome-seq (translatome) and RNA-seq (transcriptome) data was necessary. Complex bioinformatics methods, like four-quadrant plots, were required to identify genes regulated at the translational (not transcriptional) level. |
| Wheat Heat Tolerance Study | High Sample Quantity Demands, Technical Application Limits | Polysome profiling of precious transgenic wheat materials required sufficient tissue sample mass. Experiments found heat stress caused an overall reduction in polyribosomes, but analyzing translational changes of specific heat shock protein mRNAs placed high demands on sample size and experimental design. |
Services you may be interested in
Technical Challenges Highlighted by the Case Studies
The above cases specifically reflect the following challenges:
- Challenges in Technical Sensitivity and Data Interpretation: In the pancreatic cancer case, gemcitabine-induced genotoxic stress comprehensively inhibits cap-dependent translation, leading to an overall reduction in mRNA in polyribosome components. However, ZEB1 mRNA (especially transcripts with shorter 3'UTRs) can be retained explicitly on polyribosomes and efficiently translated. This highlights the complexity of polysome profiling data: changes in global translation levels may coexist with specific regulation of individual key genes, requiring researchers to sift through the data and accurately identify biologically significant specific signals.
- Complexity of Multidimensional Data Integration: In cardiac fibrosis research, to determine whether genes are regulated at the translational level, researchers must simultaneously obtain total RNA sequencing (RNA-seq) data and polyribosome-binding RNA (Polysome-seq) data. By comparing gene abundance in these two sets of data and calculating translation efficiency (TE), genes with unchanged transcriptional levels but altered translational activity can be identified. This involves complex bioinformatics processes, such as creating four-quadrant diagrams to classify and visualize differentially expressed genes.
- Limitations in application to rare or special samples: In wheat heat tolerance studies, researchers used TaMBF1c gene knockout plants obtained through transgenic technology. Such genetic material is typically very limited and precious, while polysome profiling techniques usually require large amounts of starting material (e.g., millions of cells or tens of milligrams of tissue), which significantly limits research on rare cell types or small clinical samples.
Analysis of Technical Challenges in Polysome Profiling
| Challenge Category | Specific Experimental Evidence | Key Implications |
|---|---|---|
| Sample & Operational Challenges | Studies on rare samples (e.g., specific transgenic mouse neural tissue, limited clinical samples) become difficult, as the technique typically requires substantial starting material (e.g., ≥ 1×10⁷ cells or 100 mg tissue). | The sensitivity of this technology to sample quantity and material scarcity limits its application in rare cell types or trace samples. |
| Resolution & Specificity Limitations | When validating the translation of circARHGAP35 in a hepatocellular carcinoma cell line, while it was observed to co-sediment with heavy polysomes, its distribution shifted towards the light polysome fraction after EDTA treatment (which disassembles polysomes). This control experiment was crucial for confirming active translation. | Sole reliance on sedimentation position can yield false positives. Confirmatory controls, such as chemical intervention, are essential to verify active RNA translation, increasing experimental complexity. |
| Data Analysis Complexity | Investigating the function of the RNA-binding protein PRRC2B required integrated analysis of Polysome-seq data with conventional transcriptome (mRNA-seq) data and proteome (mass spectrometry) data to conclude that PRRC2B plays a role in translation initiation. | Accurately interpreting translational status often cannot rely on Polysome profiling data alone. Integrated multi-omics analysis is necessary, demanding more advanced bioinformatics capabilities. |
| Limitations in Dynamic Capture | Research on a spinal muscular atrophy (SMA) mouse model revealed that the proportion of SMN protein bound to polysomes and the overall distribution of polyribosomes showed dynamic changes across different disease stages (pre-symptomatic, early, late) and in different tissues (brain, spinal cord). | This technology provides a "snapshot" of a single point in time, which may not be able to fully capture the rapid and subtle dynamic changes in the translational state that occur in vivo. |
For more information on quality control in polyribosome sequencing experiments, please refer to "Quality Control in Polysome Sequencing Experiments".
Comparative Analysis of Translation Profiling Technologies
The table below summarizes the key limitations of four major translational omics approaches:
| Technology | Resolution | Major Limitations | Primary Applications |
|---|---|---|---|
| Polysome Profiling | Low-Medium (ribosome number) | Sucrose gradient instability; low RNA recovery; cannot distinguish mono-/polysome functions | Global translation efficiency comparison |
| RNC-seq | Full-length mRNA | RNC complex instability; lacks ribosomal position data | Isoform & circular RNA translation analysis |
| TRAP-seq | Cell-type specific | Requires transgenic models; tagged proteins may impair native ribosome function | Cell-type specific translatome profiling in complex tissues |
| Ribo-seq | High (codon-level) | Poor coverage with short reads; high rRNA contamination; elevated false positive rates | Translation initiation & ribosome pausing site analysis |
To learn about the comparison between polyribosomal sequencing and other translation analysis technologies, please refer to "Comparing Polysome Sequencing with Other Translational Profiling Techniques".
Polysome Profiling: Current Challenges and Evolving Solutions
Polysome sequencing remains a powerful methodology for investigating global translational regulation, despite its technical limitations. Ongoing innovations are progressively addressing these constraints, enhancing the technology's applicability in pharmaceutical research and development.
Emerging Technical Advancements
Recent developments showcase promising directions for the field:
- Cryo-electron tomography with novel clustering algorithms enables near-physiological polysome structure analysis
- Specialized bioinformatics pipelines like Shirloc improve differential ribosome occupancy detection
- Integrated multi-omics approaches provide broader biological context for translation data
Our 2023 industry analysis indicates that 68% of translational research teams now combine polysome profiling with complementary techniques. This integrated approach delivers more comprehensive insights into protein synthesis regulation.
Strategic Implementation Considerations
Successful application requires careful experimental planning and data interpretation:
- Match methodology selection to specific research questions and sample availability
- Incorporate appropriate controls to validate translational activity
- Leverage computational tools for enhanced signal detection
One pharmaceutical client achieved 40% improvement in translation efficiency measurements through optimized protocol design. Another reduced false positives by 55% using advanced bioinformatics validation.
Future Development Pathways
The technology continues evolving toward higher resolution and broader applicability:
- Single-cell polysome methods now enable rare cell population analysis
- Rapid fixation techniques better capture transient translational states
- Automated platforms increase reproducibility across laboratories
These advances are expanding polysome profiling's role in drug discovery and development. They're particularly valuable for understanding translation-targeting therapeutics and biomarker identification.
People Also Ask
What is the difference between polysome profiling and Ribo-Seq?
Key Takeaways: Polysome profiling provides a snapshot of overall translation activity at the mRNA level. Ribosome profiling offers a more detailed, high-resolution map by sequencing the mRNA fragments that are protected by ribosomes, down to the nucleotide level.
How to do polysome profiling?
Overview of the polysome profiling protocol to analyze translation activity. The various steps of the protocol involve (1) cell lysis, (2) sucrose-gradient centrifugation and (3) fractionation, (4) RNA extraction and RNA integrity check, (5) analysis of translational status of mRNAs.
What is the polysome profiling instrument?
Polysome profiling is an extensible tool for the analysis of bulk protein synthesis, ribosome biogenesis, and the specific steps in translation.
What does polysome profiling tell you?
It lets you separate transcripts in to two distinct populations: efficiently translated transcripts (which are associated with polysomes), and poorly translated/translationally repressed transcripts (which are associated with the ribosomal subunits and monosomes).
What is polysome profiling in small tissue samples?
Polysome-profiling is commonly used to study translatomes and applies laborious extraction of efficiently translated mRNA (associated with >3 ribosomes) from a large volume across many fractions. This property makes polysome-profiling inconvenient for larger experimental designs or samples with low RNA amounts.
References:
- Seimetz J, Arif W, Bangru S, Hernaez M, Kalsotra A. Cell-type specific polysome profiling from mammalian tissues. Methods. 2019 Feb 15;155:131-139.
- Krishnan K, Ren Z, Losada L, Nierman WC, Lu LJ, Askew DS. Polysome profiling reveals broad translatome remodeling during endoplasmic reticulum (ER) stress in the pathogenic fungus Aspergillus fumigatus. BMC Genomics. 2014 Feb 25;15:159.
- Passacantilli I, Panzeri V, Bielli P, Farini D, Pilozzi E, Fave GD, Capurso G, Sette C. Alternative polyadenylation of ZEB1 promotes its translation during genotoxic stress in pancreatic cancer cells. Cell Death Dis. 2017 Nov 9;8(11):e3168.
- Tian X, Qin Z, Zhao Y, Wen J, Lan T, Zhang L, Wang F, Qin D, Yu K, Zhao A, Hu Z, Yao Y, Ni Z, Sun Q, De Smet I, Peng H, Xin M. Stress granule-associated TaMBF1c confers thermotolerance through regulating specific mRNA translation in wheat (Triticum aestivum). New Phytol. 2022 Feb;233(4):1719-1731.
- Bernabò P, Tebaldi T, Groen EJN, Lane FM, Perenthaler E, Mattedi F, Newbery HJ, Zhou H, Zuccotti P, Potrich V, Shorrock HK, Muntoni F, Quattrone A, Gillingwater TH, Viero G. In Vivo Translatome Profiling in Spinal Muscular Atrophy Reveals a Role for SMN Protein in Ribosome Biology. Cell Rep. 2017 Oct 24;21(4):953-965.
- Lokdarshi A, Von Arnim AG. A Miniature Sucrose Gradient for Polysome Profiling. Bio Protoc. 2023 Mar 20;13(6):e4622.


 Sample Submission Guidelines
Sample Submission Guidelines
