Phage E. coli Sequencing: Applications and Considerations
Phage E. E. coli sequencing is an important tool to study the interaction between bacteriophages and host bacteria (E. coli) . With the rapid development of genomics technology, Phage E. E. coli sequencing not only helps scientists to understand the genetic characteristics of bacteriophages, but also provides a new research perspective for biotechnology, medicine and environmental protection. This article will discuss the bacteriophage E. Applications, potential challenges, and considerations of E. coli sequencing.
Core Application
Genomic Targeting of Drug-Resistant Pathogens
Mechanistic Insights from E. coli O157 Phages
- Group-Specific Infection Patterns: Phage T4 isolates formed three distinct genomic clusters. Each cluster exhibited preferential infection of specific PT strains (e.g., Group 1 phages consistently lysed PT21/28 variants).
- Phenotype-Genotype Correlation: Infection profiles within clusters showed >85% similarity, with genomic divergence <5% between group members. Group 1 possessed five unique genes potentially governing host specificity.
- Cocktail Design Implications
- This clustering enables rational phage cocktail development:
- Universal components: Broadly active phages across clusters
- Specific components: Group-targeting phages for resistant strains
- This clustering enables rational phage cocktail development:
- Therapeutic Targeting Strategy: Minimal genetic variations between clusters (particularly in tail fiber genes) represent precise engineering targets for enhanced host range control (Cowley LA et al., 2015).
Therapeutic Candidate Validation: VE20 Phage Case
Genomic Safety Profile
| Risk Factor | VE20 Status | Validation Method |
|---|---|---|
| Antibiotic resistance | Absent (no bla, aac genes) | Systematic ORF screening |
| Virulence factors | No Shiga toxin (stx) | Homology analysis |
| Lysogenic potential | No integrase (int) | Module detection |
Environmental Safety Assurance
- <75% homology to resistance-gene carriers (e.g., sewage isolate YZ1 with SUL2)
- Minimal horizontal gene transfer risk
Molecular Determinants of Therapeutic Efficacy
Host Range Expansion
- Tail filament variations (ORF13-14) enable lysis of:
- 47.05% clinical E. coli strains
- Salmonella enterica serovars
Replication Efficiency
- Short latency (10 min): Driven by DNA replication genes (ORF72-73)
- Moderate burst size (60 PFU/cell): Streamlined lysis module (single endolysin ORF16)
Environmental Resilience
- Thermostability (30-60°C): Capsid structural integrity
- pH tolerance (3-11): Virion envelope adaptations (Zhong Z et al., 2024)
 The lysis rate of phage vE20 in different MOI (Zhong Z et al., 2024)
The lysis rate of phage vE20 in different MOI (Zhong Z et al., 2024)
Novel Myoviridae Phage VB: Characterization and Therapeutic Potential
Isolation and Host Specificity
Phage VB (GenBank: MT664721), a newly characterized member of the Myoviridae family, demonstrates direct lytic activity against multidrug-resistant Escherichia coli APEC O78. This pathogen represents a major avian disease agent with significant zoonotic transmission potential.
Therapeutic Applications
- Targeted MDR Intervention: Precision therapy against drug-resistant APEC O78 infections in poultry and humans
- Extended Antimicrobial Spectrum
- Preliminary efficacy against critical nosocomial pathogens:
- Acinetobacter baumannii
- Pseudomonas aeruginosa (Deng S et al., 2021)
- Extended Antimicrobial Spectrum
Synthetic Biology
This study successfully engineered a fluorescent phage, K1F-GFP, targeting pathogenic Escherichia coli K1 using CRISPR/Cas technology. We provide the first mechanistic insights into its efficient clearance of intracellular pathogens within human cells.
1. Phage Engineering Breakthroughs
- Targeted Gene Insertion: Employing CRISPR/Cas-assisted homologous recombination, the GFP gene was precisely integrated into a non-essential region of the phage genome, yielding a stable recombinant.
- Functional Capsid Optimization: Defects associated with capsid protein fusion (e.g., reduced fitness from G10::gfp) were overcome by optimizing linker peptide design, enabling functional fluorescent protein expression.
2. Intracellular Bactericidal Mechanism
- Co-invasion Pathway: Phage K1F-GFP co-enters T24 human bladder epithelial cells via phagocytosis alongside its host bacterium (E. coli EV36-RFP), confirmed by co-localization with the Rab7 phagosomal marker.
- Enhanced Intracellular Killing: K1F-GFP significantly reduces intracellular bacterial load, demonstrating >50% higher bactericidal efficiency within cells compared to the extracellular environment. Specificity was confirmed by the inactivity of T7 phage and against non-sensitive bacteria.
- Degradation Mechanisms:
- Phage-Bacteria Complexes: Degraded via LC3-associated phagocytosis (validated by co-localization with the lysosomal marker Cathepsin L).
- Free Phages: Subject to a novel xenophagic clearance pathway, independent of the ubiquitin-NDP52 autophagy route (Møller-Olsen C et al., 2018).
Oral Microencapsulated Phage A221: Efficacy Against Piglet Diarrhea
Phage Characterization
- Classification: Ackermannviridae family (Aglimvirinae subfamily, Agtrevirus genus)
- Genome: 153,297 bp dsDNA
- Host Specificity: Targets Escherichia coli GXXW-1103
- In Vitro Efficacy: Potent antibacterial activity (≥16 hr sustained suppression)
Microencapsulation Technology
- Core Innovation: Gastric acid-resistant formulation protects phage viability
- Delivery Mechanism: Targeted intestinal release
In Vivo Therapeutic Outcomes
| Parameter | Results | Significance |
|---|---|---|
| Daily Weight Gain | Significantly increased (p<0.001) | Enhanced growth performance |
| Enterobacteriaceae Abundance | Reduced to 0.64% | Pathogen suppression |
| Bacterial Load Reduction | ||
| - Jejunal lymph nodes | Marked reduction (p<0.001) | Systemic protection |
| - Cecum & spleen | Dramatically lowered (p<0.001) | Tissue colonization control |
| Intestinal Pathology | Mucosal injury & inflammation alleviated | Structural/functional recovery |
Therapeutic Benchmark
Achieved equivalent efficacy to antibiotic florfenicol
Translational Impact
- Antibiotic Alternative: Effective against MDR-induced diarrhea
- Sustainable Agriculture: Enables green transition in livestock production (Mao X wt al., 2023)
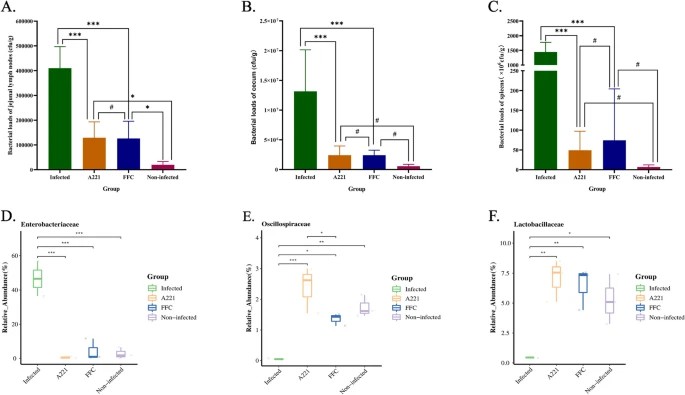 Effects of phage A221 or FFC on bacteria (Mao X wt al., 2023)
Effects of phage A221 or FFC on bacteria (Mao X wt al., 2023)
Phage Sequencing Workflow: Technical Framework
1. Sample Processing
- Phage Enrichment
- Virus concentration: PEG8000 precipitation
- Purification: Iodixanol density gradient centrifugation
- Host DNA Removal
- Enzymatic digestion: DNase I treatment
- Bioinformatics filtration: Alignment against E. coli K-12 genome
2. Nucleic Acid Preparation
- DNA Extraction
- Capsid lysis: CTAB method
- Purification: Silica membrane column chromatography
3. Sequencing Strategy
- Dual-Platform Approach
- Short-read: Illumina PE150
- Long-read: Oxford Nanopore PromethION
4. Hybrid Assembly & Polishing
- Input Data Preparation:
- Illumina short-read sequencing data serves as input for a de novo assembly using SPAdes.
- Nanopore long-read sequencing data serves as input for a de novo assembly using Flye.
- Hybrid Assembly Polishing:
- The assembled contigs from both SPAdes (Illumina-based) and Flye (Nanopore-based) are combined.
- These contigs undergo iterative polishing with Racon to improve accuracy and resolve structural errors.
- Final Output:
- The polished assembly yields high-quality contigs with a minimum length of >100 kilobases (kb), achieving an N50 statistic (a metric indicating half of the assembly is contained in contigs of this size or larger).
- Key Technologies:
- SPAdes: Optimized for Illumina short reads.
- Flye: Designed for noisy long reads (e.g., Nanopore).
- Racon: A consensus tool that refines assemblies using both short and long reads.
5. Functional Annotation
- Dual-Database Screening
- PHROGs: Core phage functions (replication, packaging, structure)
- VFDB: Virulence factors (e.g., Shiga toxin) & resistance genes (e.g., β-lactamases)
Key Technical Optimizations
| Challenge | Solution | Improvement |
|---|---|---|
| Low nucleic acid recovery (<40%) | Ultrafiltration + iodixanol gradient | ↑72% yield |
| Assembly fragmentation | Hybrid long-short read assembly | N50 >100kb |
| Low host prediction accuracy (<60%) | VHM-Net machine learning model | 89% accuracy |
| Lysogeny detection gaps | Integrated int/ci/attP site scanning | Comprehensive identification |
For a more detailed approach to phage sequencing, please refer to "Phage Genome Sequencing: Methods, Challenges, and Applications".
For more information on what phage sequencing is, see "What Is Phage Sequencing? A Complete Guide for Researchers".
For more information on the protocol for Lambda Genome Sequencing, please refer to "Lambda Phage Genome Sequencing: Protocols and Use Cases".
Key Considerations
1. Control of Biological Characteristics
- Mandatory Virulence Gene Screening: Bacteriophages encoding Shiga toxin (STX1/2) or enterotoxin (AstA) must be excluded from use, regardless of the gene's functional status (e.g., inactivation).
- Industrial Strain Suitability: Phages intended for industrial applications require careful assessment to eliminate lysogenic variants (e.g., Lambda *cI+*). This prevents potential horizontal gene transfer events during fermentation processes.
2. Dynamic Monitoring of Host-Range Variation
A periodic assessment strategy, implemented every 10 passages of continuous culture, is essential:
- Host-Range Validation via Spot Assay: Detect alterations in phage lysis spectra using a standardized host strain library, employing the dot (spot) assay method. Quantify changes in lytic plaque diameter and clarity to evaluate host-range excursions.
- Genome-Wide Mutation Analysis:
- Perform whole-genome sequencing(WGS) on the current phage generation.
- Align sequences to the initial reference genome (e.g., using BWA-MEM + Samtools) to identify variants, including:
- Single nucleotide polymorphisms (SNPs) and insertion/deletion events (Indels).
- Structural variations (e.g., gene inversions, repeat amplifications).
- Recombination events (visualized via circular genome alignments in tools like BRIG).
3. Technical Implementation
- Sequencing Depth and Quality Control:
- Basic Research: Minimum coverage ≥30x; Q30 score > 90%.
- Clinical/Industrial Applications: Minimum coverage ≥50x; requires triple testing with manual sequence review.
- Contamination Control:
- Filter all reagents through a 0.22 μm membrane.
- Include no-template controls (NTCs) in all procedures to detect and prevent cross-contamination.
4. Safety and Compliance Requirements
- Biosafety Protocols:
- Handle phages carrying virulence factors (e.g., STX) within Biosafety Level 2 (BSL-2) laboratories.
- Environmental release of engineered phages necessitates triple physical containment barriers, such as microfluidic closed systems.
- Intellectual Property Management:
- Secure patents for core components (e.g., the JS98 phage tail fiber protein GP37 structure).
- Deposit engineered phage strains according to the Budapest Treaty in recognized international repositories.
Explore our Service →
Directions for Cutting-Edge Breakthroughs
1. Real-Time In Situ Monitoring
Implement direct nanopore sequencing of intestinal mucus samples, achieving single-cell resolution without prior enrichment. This enables real-time observation of critical interactions, such as those between phages and Clostridioides difficile.
2. Evolutionary Blocking of Resistance
Develop CRISPR-Cas9-assisted phages. These carry sgRNAs designed to target essential host resistance genes (e.g., MEFA). This strategy achieves high-efficiency inactivation (>98%) of targeted resistance mechanisms.
3. Microfluidic High-Throughput Platform
Establish integrated microfluidic systems combining: single-phage sorting via fluorescent labeling, 96-channel parallel sequencing capacity, and real-time lysis dynamic imaging. This platform significantly accelerates the screening pipeline for identifying potent virulent phages.
Implementation Considerations
Design Phase
- Host Bacterium Status: Optimize infection efficiency by utilizing E. coli hosts in the logarithmic growth phase (OD600 = 0.4 ± 0.05).
- Sample Representativeness: For environmental samples, employ multi-point collection strategies (e.g., across different locations within a fermenter) to minimize sampling bias.
Data Analysis Phase
- Decontamination: Mandatorily align sequence data against the E. coli K-12 MG1655 reference genome to identify and remove contaminating sequences.
- Contamination Control: Rigorously apply negative controls throughout the workflow to detect and mitigate potential cross-contamination issues.
Conclusion
Phage-E. coli sequencing technology holds transformative potential across medical targeted therapy, food safety surveillance, and synthetic biology applications. Its successful deployment necessitates simultaneous attention to technical precision (hybrid assembly, sufficient sequencing depth), stringent biosafety protocols (e.g., dual-review for virulence genes), and application-specific suitability (e.g., lysogen exclusion in industrial settings). The integration of nanopore real-time sequencing with CRISPR-based dynamic editing is poised to accelerate breakthroughs in overcoming drug-resistant bacteria, fundamentally reshaping the landscape of precision microbial engineering.
References:
- Cowley LA, Beckett SJ, Chase-Topping M, Perry N, Dallman TJ, Gally DL, Jenkins C. Analysis of whole genome sequencing for the Escherichia coli O157:H7 typing phages. BMC Genomics. 2015 Apr 8;16(1):271.
- Zhong Z, Wang Y, Li H, Zhang H, Zhou Y, Wang R, Bao H. Characterization and genomic analysis of a novel E. coli lytic phage with extended lytic activity against S. Enteridis and S. Typhimurium. Food Prod Process and Nutr. 6, 14 (2024).
- Deng S, Xu Q, Fu Y, Liang L, Wu Y, Peng F, Gao M. Genomic Analysis of a Novel Phage Infecting the Turkey Pathogen Escherichia coli APEC O78 and Its Endolysin Activity. Viruses. 2021 May 31;13(6):1034.
- Møller-Olsen C, Ho SFS, Shukla RD, Feher T, Sagona AP. Engineered K1F bacteriophages kill intracellular Escherichia coli K1 in human epithelial cells. Sci Rep. 2018 Dec 3;8(1):17559.
- Mao X, Wu Y, Ma R, Li L, Wang L, Tan Y, Li Z, Liu H, Han K, Cao Y, Li Y, Peng H, Li X, Hu C, Wang X. Oral phage therapy with microencapsulated phage A221 against Escherichia coli infections in weaned piglets. BMC Vet Res. 2023 Sep 20;19(1):165.


 Sample Submission Guidelines
Sample Submission Guidelines
