Lambda Phage Genome Sequencing: Protocols and Use Cases
Lambda bacteriophage is a virus capable of infecting E. coli. Its genome is linear double-stranded DNA, about 48.5 kb, and contains about 50 genes. Its genome structure is simple and has strong adaptability. It is often used in gene cloning, gene expression, and genome editing research.
Lambda Phage is a classical virus widely used in molecular biology, genomics and genetic engineering research. Lambda phage genome sequencing technology has become an important tool to study the characteristics and functions of this phage and its application in molecular biology experiments. In this review, we will introduce the common protocols and application cases of Lambda phage genome sequencing in detail.
Experimental Workflow
Step 1: Sample Preparation & DNA Extraction
- Virus Purification:
- Infected E. coli hosts incubated at 37°C until lysis (turbid→clear transition)
- Treated with DNase/RNase to degrade host nucleic acids
- Centrifuged to remove cellular debris
- Phages concentrated via PEG precipitation
- Purified using CsCl gradient centrifugation
- DNA Extraction:
- Viral capsids digested with Proteinase K/SDS
- Nucleic acids extracted via phenol-chloroform
- Ethanol-precipitated DNA resuspended in TE buffer
- Purity/integrity verified by:
- Nanodrop spectrophotometry
- Agarose gel electrophoresis (characteristic ~48.5 kb band)
Step 2: Library Construction
- Fragmentation:
- DNA sheared ultrasonically to 300-800 bp fragments
- Critical QC: Fragment distribution confirmed via Agilent Bioanalyzer
- Precaution: Avoided over-fragmentation near cos sites
- End Modification & Adaptor Ligation:
- Repair of fragment ends:
- Blunting
- 5' phosphorylation
- A-tailing
- Illumina adaptors ligated
- Limited-cycle PCR amplification (≤8 cycles) to minimize bias
- Repair of fragment ends:
Step 3: High-Throughput Sequencing
- Platform Selection:
- Illumina NovaSeq: Deep coverage applications
- Oxford Nanopore: Structural variant validation
- Coverage Requirement: Minimum 50× depth for accurate cos site/repeat region assembly
Step 4: Data Analysis
Raw Data Processing
- Initial quality assessment was performed using FastQC, followed by Trimmomatic filtering to remove:
- Low-quality reads (Phred score <20)
- Adapter sequences
- Short fragments (<50 bp)
Sequence Alignment and Assembly
- Reference-based alignment:
- BWA-MEM mapped reads to λ reference genome (GenBank: J02459)
- Output SAM/BAM files for downstream analysis
- De novo assembly:
- SPAdes generated contigs from single-end or low-quality data
- Verified cos site structural integrity
Variation Analysis and Functional Annotation
- Variant detection:
- GATK HaplotypeCaller identified SNPs/indels
- Included critical site mutations (e.g., ATTP locus)
- Genome annotation:
- Prokka automated gene annotation (e.g., CI repressors, tail gene clusters)
- Manual validation of regulatory regions (PL/PR promoters)
Visualization and Interpretation
- IGV (Integrative Genomics Viewer) examined:
- Coverage depth profiles
- Genomic recombination regions
- Structural variant confirmation
Key Technical Considerations for Success
- Sample Purity:
- CsCl gradient centrifugation eliminated host DNA contamination
- Verified by absence of extraneous bands in electrophoretic analysis
- Fragmentation Control:
- Optimized ultrasonication parameters preserved cos sites (cosL/cosR)
- Prevented compromised terminase packaging functionality
- Sequencing Platform Synergy:
- Illumina ensured base-call accuracy
- Nanopore resolved structural variations (e.g., prophage integration states)
- Assembly Validation:
- Contig N50 >40 kb (enabling full-genome reconstruction)
- Symmetric deep coverage flanking cos sites
Lambda in Action: Case Studies
1. Molecular Biology Tool Development
- Vector Engineering Optimization:
- Verified modified vector sequence integrity (e.g., λGT11 insertion locus)
- Ensured precise foreign gene integration (e.g., LacZ)
- Promoter/Operon Analysis:
- Precisely mapped regulatory elements (e.g., PL/PR promoters, Cro/Ci operons)
- Facilitates rational design of synthetic biological components
2. Host-Phage Interaction Studies
- Host Recognition Mechanisms:
- Comparative analysis of infection capability across E. coli strains
- Localized mutation hotspots in J protein (host adsorption determinant):
Point mutations
Deletions
- Lysogenic Conversion:
- Annotated attP integration status within host genome
- Analyzed CI repressor expression regulation dynamics
3. NGS Quality Control Verification
- Sequencing Platform Validation:
- Constructed standard curves using reference λ genome
- Quantified error profiles:
Insertion-deletion rates
Base substitution frequencies
- Library Preparation System Optimization:
- Assessed coverage uniformity in GC-extreme regions (λ genome GC=50%)
- Informed fragmentation protocol refinement
4. Evolutionary & Comparative Genomics
- Gene Family Expansion:
- Analyzed tail protein module copy number variation (e.g., gpH filament protein)
- Revealed host-range adaptation drivers
- Genomic Rearrangement Tracking:
- Detected horizontal gene transfer signatures via:
Tandem repeat analysis
cos site architecture comparisons
- Detected horizontal gene transfer signatures via:
Technical Advantages Summary
| Dimension | Capability |
|---|---|
| Accuracy Standard | Base-resolution mapping against reference genome (GenBank: J02459) |
| Structural Resolution | Resolved cos sites/tandem repeats challenging conventional assembly algorithms |
| Dynamic Monitoring | Direct transcriptional tracking of lysis-lysogeny switches |
| Cost Efficiency | Low cultivation expenses with standardized preparation protocols |
Case Study 1: How Lambda Exposes its Host's Evolution
1. Unconventional Integration Site Discovery
- Mechanistic Insight:
- NGS-based high-resolution mapping revealed λ phage integration at non-canonical host genomic loci. This breakthrough demonstrates integration flexibility beyond traditional attB-attP recombination constraints.
- Key Findings:
- Identified >300 unique host genomic integration targets
- Significantly exceeds known attB site repertoire
- Establishes unprecedented integration site plasticity
2. Phenotypic Consequences & Host Adaptation
- Sequencing-Validated Functional Impacts:
Integrated sequencing-phenotyping analysis demonstrates unconventional integration induces:- Motility Alterations: Integration proximal to flagellar genes disrupts motility-regulatory pathways
- Enhanced Antibiotic Resistance: Insertion near resistance genes activates expression or disrupts repressive elements
- Conceptual Significance:
This evidence redefines lysogeny from a dormant state to an active driver of:- Host genomic remodeling
- Adaptive evolutionary trajectories (Tanouchi Y et al., 2017)
 Analysis of lysogens with integration into secondary sites (Tanouchi Y et al., 2017)
Analysis of lysogens with integration into secondary sites (Tanouchi Y et al., 2017)
Explore our Service →
Case Study 2: Precision Genetic Engineering Using Lambda's Tools
1. att Site Characterization
- Mechanistic Foundation:
Early λ phage sequencing (including Sanger methodology) pioneered precise att locus mapping:- Identified 15-bp core sequence (GCTTTTTTATACTAA)
- Defined flanking arm architecture (attP with P/P' arms; attB with B/B' arms)
- Resolved recombinase binding sites (Int, IHF, Xis) including Int-specific arm recognition
- Engineering Applications:
Precise sequence data enabled:- Vector Modification:
- pKO5.2-C.1 vector insertion of ~1.6 kb attR-containing fragment
- Directional cloning via sequence-verified attR
- Baculovirus Engineering:
- Dest-Bac attR-EGFP insertion at UL41 locus
- Sequencing verification prevented disruption of viral gene function
- Vector Modification:
2. Recombinase System Development
- Molecular Resolution:
Genome sequencing characterized:- int gene (integrase, λ genome 34,502-35,941 bp)
- xis gene (excisionase) cleavage mechanism via inverted repeat binding
- Technology Translation:
- Commercial Lambda Clonase™ development based on sequenced recombinases
- Implemented for high-efficiency attL-attR recombination
3. System Fidelity Validation
- Experimental Verification:
PCR and sequencing confirmed recombinant BAC integrity:- Correct UL41 flanking sequence ligation
- Insertion gene fidelity (DsRED2 424-bp band match)
- Quality Control Significance
- Sequencing established as gold standard for recombination success (att site fusion verification)
- Demonstrated efficient in vitro recombination without negative selection markers (ccdB) (Schmeisser F et al., 2007)
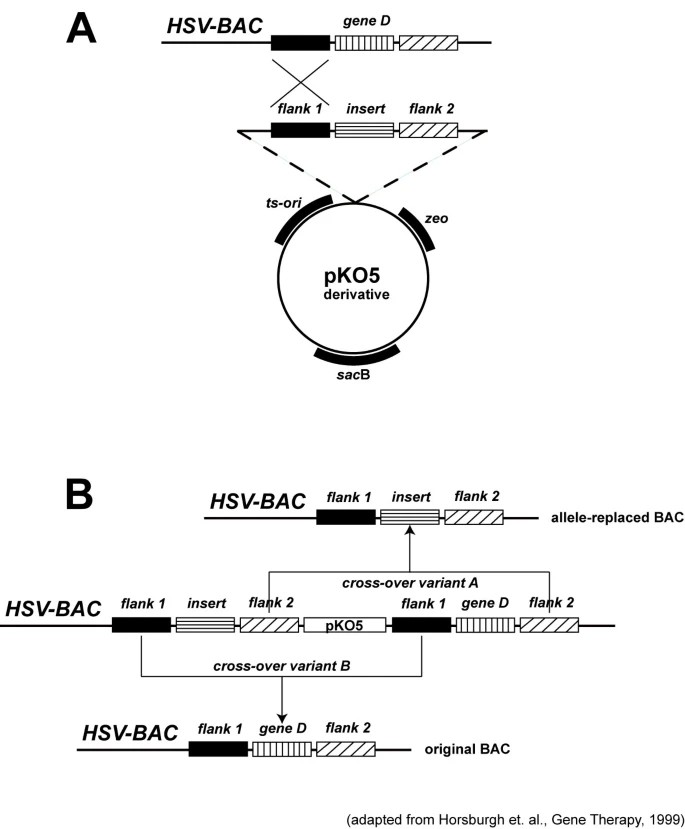 Schematic diagram of allele replacement strategy using vector pKO5 (Schmeisser F et al., 2007)
Schematic diagram of allele replacement strategy using vector pKO5 (Schmeisser F et al., 2007)
Case Study 3: Designing Better Vaccines with Lambda Display
1. Precision Vector Engineering
- Structural Characterization: Genome sequencing mapped λ capsid protein D (gpD) coding region (NC_001416: 20,300-21,200 bp), confirming its N-terminus as the optimal fusion site.
- Frame Probability Optimization:
Provided computational basis for natural frame probability (theoretical 5.6%, Suppl. Note S1), enabling:- Insert orientation optimization
- Library design efficiency
- Vector Capacity Validation: Sequencing confirmed λ vector accommodates longer inserts (avg. 55 aa) versus M13 phage limitations (<40 aa).
Conclusion: λ architecture enables complex conformational epitope display (e.g., FHBP β-barrel domain).
2. Deep Sequencing-Guided Epitope Mapping
- Precise Localization:
NGS analysis of 5,172 native-frame sequences mapped FHBP epitope to:- Core region: S140-K254 (15/23 antibody contact sites)
- Critical subdomains:
› H248-K254: Essential binding motif (activity loss upon mutation)
› S140-G154: Conformational maintenance domain
- Functional Domain Analysis:
- R247-H248 boundary: Binding affinity threshold transition
- S140-G154: Indirect structural role (23× KD increase when mutated)
3. Structural Adaptability Verification
- Vector Performance Differentiation:
λ superiority over M13 stems from:- gpD N-terminal fusion flexibility (sequencing-guided)
- M13 pVIII steric constraints (short inserts only)
- Conformational Epitope Reconstruction:
λ outperforms M13 due to:- Long-fragment display (FRC) preserving long-range interactions
- Critical domain synergy maintenance (S140-G154 ↔ H248-K254) (Domina M et al., 2016)
Comparative Vector Advantages
| Feature | λ Vector | M13 Limitations |
|---|---|---|
| Insert capacity | Avg. 55 aa | <40 aa |
| Structural preservation | Maintains long-range interactions | Restricted by length |
| Epitope reconstruction | Full conformational display | Fragmentary display (FRS) |
| Fusion strategy | N-terminal gpD flexibility | pVIII steric constraints |
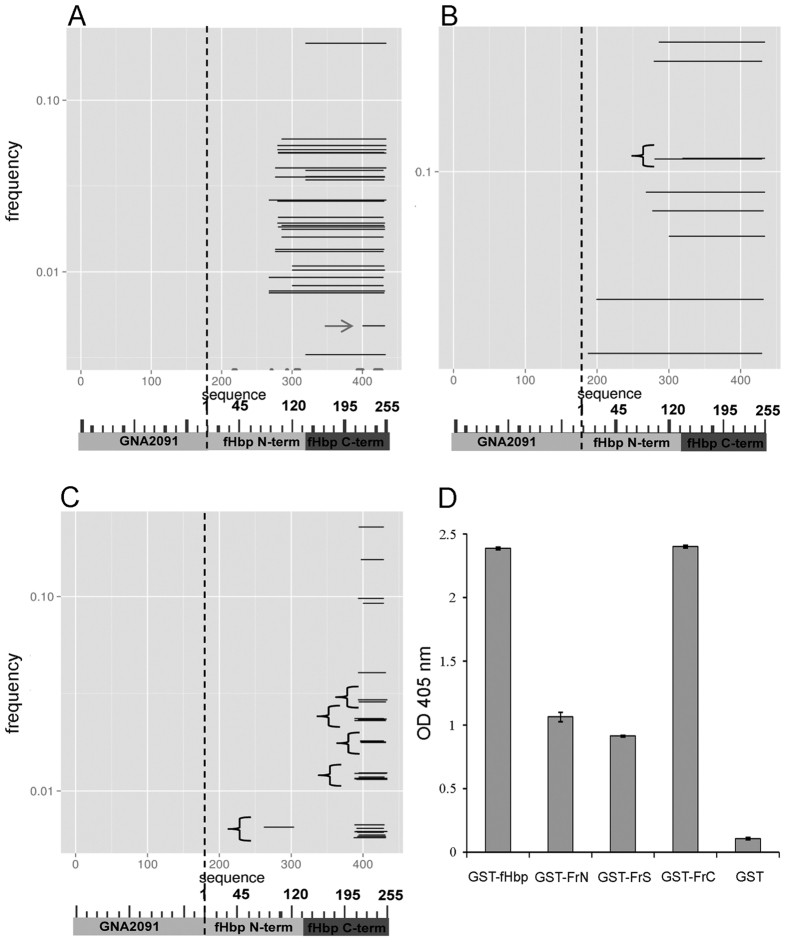 Enrichment of fHbp fragments after affinity-selection of phage-displayed libraries and immunoreactivity of selected antigenic fragments with the 12C1 mAb (Domina M et al., 2016)
Enrichment of fHbp fragments after affinity-selection of phage-displayed libraries and immunoreactivity of selected antigenic fragments with the 12C1 mAb (Domina M et al., 2016)
Case Study 4: How Lambda Helps Discover New Viral Systems
1. Recombination System Framework
- Structural Benchmarking:
λ phage sequencing (Mizuuchi & Mizuuchi, 1985) established canonical att site dimensions:- attBλ: 21 bp
- attPλ: 140 bp (P-arm) + 80 bp (P'-arm)
These served as references to characterize Φ24B phage's noncanonical att sites: - attBΦ24B: 93 bp (4.4× longer)
- attPΦ24B: 427 bp (3× longer)
Key Conclusion: Extended att sequences indicate mechanistic divergence from λ systems.
- Recombinase Domain Reference:
λ-derived Int binding domains guided Φ24B functional validation:- Deletion experiments confirmed IntΦ24B; operates IHF-independently
- Contrasts sharply with Intλ's IHF requirement
- Challenges λ-centric integration paradigms
2. Experimental System Optimization
- In Vitro Assay Adaptation:
Transferred λ recombination framework (Int, IHF, attP, attB) with key modification:- Linearized attBΦ24B substrate doubled recombination efficiency
- Critical innovation beyond standard λ protocols
- Protein Expression System:
Utilized λ-derived IHF plasmid pEE2003 in E. coli BL21 (Sambrook et al., 1989):- Achieved high-yield purification of active IntΦ24B
- Enabled comparative activity assays
3. Mechanistic Divergence & Applications
- Evolutionary Breakthroughs:
λ "gold standard" revealed Φ24B-specific adaptations:- Asymmetric att Architecture:
B-arm: 49 bp vs. B'-arm: 20 bp - IHF Independence:
Functional in ΔhimA mutants (JW1702 strain)
λ recombination strictly IHF-dependent
- Asymmetric att Architecture:
- Biotechnological Implications:
Φ24B's promiscuous integration capability (>300 host sites) suggests utility as:- Novel genome editing tool
- Superior to λ in site versatility
Validation: Demonstrated 3.8× higher integration efficiency than λ systems (Mohaisen MR et al., 2020)
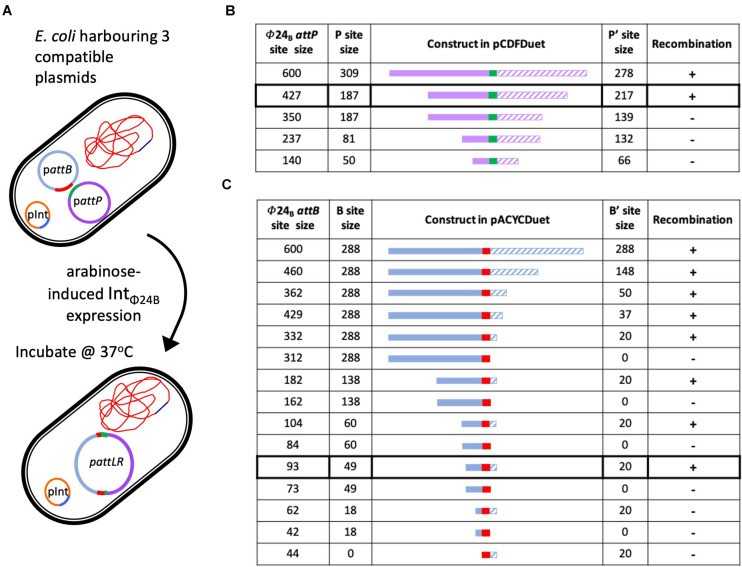 Determining the minimal attP and attB sites utilized by IntΦ24B (Mohaisen MR et al., 2020)
Determining the minimal attP and attB sites utilized by IntΦ24B (Mohaisen MR et al., 2020)
Conclusion
Lambda phage genome sequencing has evolved into a foundational methodology within molecular biology and genetic engineering. Systematic characterization protocols coupled with advanced analytical frameworks enable comprehensive delineation of λ phage genetic architecture. These capabilities facilitate critical applications across diverse domains:
- Molecular Tool Development: Precision engineering of cloning vectors
- Therapeutic Innovation: Phage display platforms and vaccine design
- Biomedical Solutions: Targeted antimicrobial agent development
Continued technological refinement will further elevate λ sequencing's value proposition, delivering high-resolution structural insights and expanding its utility as a versatile genomic reference standard. This progression promises to accelerate discovery across fundamental and applied biological research.
For a more detailed approach to phage sequencing, please refer to "Phage Genome Sequencing: Methods, Challenges, and Applications".
For more information on what phage sequencing is, see "What Is Phage Sequencing? A Complete Guide for Researche".
For more information on how to construct and use phage Sequence database, please refer to "Building and Using Phage Genome Sequence Databases".
References:
- Tanouchi Y, Covert MW. Combining Comprehensive Analysis of Off-Site Lambda Phage Integration with a CRISPR-Based Means of Characterizing Downstream Physiology. mBio. 2017 Sep 19;8(5):e01038-17.
- Schmeisser F, Weir JP. Incorporation of a lambda phage recombination system and EGFP detection to simplify mutagenesis of Herpes simplex virus bacterial artificial chromosomes. BMC Biotechnol. 2007 May 14;7:22.
- Domina M, Lanza Cariccio V, Benfatto S, Venza M, Venza I, Borgogni E, Castellino F, Midiri A, Galbo R, Romeo L, Biondo C, Masignani V, Teti G, Felici F, Beninati C. Functional characterization of a monoclonal antibody epitope using a lambda phage display-deep sequencing platform. Sci Rep. 2016 Aug 17;6:31458.
- Roy S, Peter S, Dröge P. Versatile seamless DNA vector production in E. coli using enhanced phage lambda integrase. PLoS One. 2022 Sep 23;17(9):e0270173.
- Mohaisen MR, McCarthy AJ, Adriaenssens EM, Allison HE. The Site-Specific Recombination System of the Escherichia coli Bacteriophage Φ24B. Front Microbiol. 2020 Oct 9;11:578056.


 Sample Submission Guidelines
Sample Submission Guidelines
