Case Study: Erwinia Phage Sequencing for Agricultural Applications
Plant bacterial diseases represent a major limiting factor in global agricultural productivity. Pathogens such as Erwinia spp., which cause soft rot, persistently threaten numerous high-value crops. The longstanding overreliance on chemical pesticides has led to significant challenges, including environmental contamination and the emergence of resistant strains. Consequently, developing sustainable and efficient alternative control strategies has become an urgent priority.
Within this context, bacteriophages—viruses capable of specifically targeting and lysing pathogenic bacteria—have gained attention as promising biocontrol agents, owing to their high specificity and environmental compatibility. However, the successful field application of phage-based solutions necessitates thorough verification of their safety and efficacy, which in turn requires a detailed understanding of their genetic makeup.
This case study systematically demonstrates the use of high-throughput sequencing for comprehensive genomic characterization of Erwinia-targeting phages with biocontrol potential. By outlining the molecular and bioinformatic methodologies employed to transition a raw phage isolate into a well-characterized biocontrol candidate, this work offers a reproducible technical pathway and evaluation framework. It aims to support standardization and commercial development in the field of agricultural phage applications.
Case 1: 1. Phage Sequencing: Unlocking the Genomic Blueprint for Next-Generation Biocontrol
Confronted with the destructive impact of fire blight caused by Owen amyloliquefaciens, conventional chemical control methods are increasingly challenged by environmental concerns and issues of drug resistance. Phage-based biological control presents a promising targeted alternative; however, its practical implementation has long been hindered by uncertainties surrounding host range and environmental stability. In a groundbreaking approach, Sabri M et al. employed high-throughput phage sequencing technology to precisely decipher the genomic architecture of virulent phages, thereby fundamentally elucidating their host specificity and lytic mechanisms. This research aims to develop novel, highly efficient and targeted bio-pesticides, ultimately providing a powerful and eco-friendly intelligent control strategy for sustainable agriculture.
Emergence of Phage Biocontrol
Mounting demands for sustainable agriculture have accelerated interest in bacteriophage-mediated biological control. As natural bacterial predators, phages offer targeted pathogen elimination through specific infection and lysis mechanisms. Their inherent advantages include precise host specificity, environmental compatibility, and potential for resistance mitigation.
Implementation Challenges and Technological Solutions
Despite promising attributes, practical phage deployment faces three primary constraints:
- Variable host strain specificity
- Limited environmental persistence
Incompletely characterized lysis systems
These factors collectively restrict field efficacy. Crucially, advances in high-throughput sequencing now enable comprehensive analysis of phage genomic architecture, host recognition pathways, and lytic machinery - substantially accelerating biopesticide development.
Study Rationale and Objectives
This investigation bridges fundamental and applied research by integrating multi-omics approaches to systematically characterize an Erwinia-targeting phage. Through concurrent analysis of genomic features, lysis mechanisms, and field performance, we establish a scientific foundation for agricultural implementation of phage biocontrol against fire blight.
2. Materials and Methods
2.1 Phage Isolation and Purification
Phages were isolated from infected orchard soil/water samples using host-dependent enrichment. The protocol comprised:
- Host Preparation: Erwinia amylovora strain PGL Z1 served as the host
- Enrichment: 10 g soil samples incubated in TSB medium (25°C, 24-48 h, 180 rpm)
- Clarification: Supernatant centrifugation (8,000 × g, 10 min) followed by 0.45 μm filtration
- Plaque Assay: Double-layer agar method (0.35% top agar optimized for giant phage recovery)
- Purification: Three successive plaque-to-plaque isolations yielded monoclonal phages
2.2 Genomic Sequencing and Annotation
- DNA Processing:
- Extraction: Norgen Biotek phage-specific kit
- Library Prep: NEB Ultra II FS DNA kit
- Sequencing: Illumina MiSeq (2×250 bp) or PacBio platforms
- Bioinformatic Analysis:
- Assembly: De novo assembly via SPAdes v3.12
- ORF Prediction: GeneMarkS v4.25 with BLASTP validation (e-value ≤0.001)
- Functional Annotation: KEGG, COG, and VFDB databases
- Phylogenetics: Maximum-likelihood tree (major capsid protein)
- Packaging Analysis: Terminal structure prediction using PhageTerm
2.3 Lysis Mechanism Validation
Key lysis genes (holin, endolysin, spanin) were functionally characterized:
- Cloning: pET-vector expression constructs
- Lysis Kinetics: Transformant viability curves in E. coli BL21(DE3)
- Secretion Analysis: Sodium azide-based SEC pathway inhibition
- Signal Peptide Mutagenesis: N-terminal deletion mutant assays
2.4 Efficacy Assessment
- Plant Models:
- Greenhouse: 1-year-old 'Conference' pear saplings
- Field: Mature wheat (Triticum aestivum cv. AAI-W6)
- Treatment Protocol:
- Stem inoculation with E. amylovora (~10⁸ CFU/mL)
- Phage suspension application (~10⁸ PFU/mL, MOI=1) after 30 min
- Controls: Pathogen-only (positive), PBS (negative), streptomycin (100 μg/mL)
- Monitoring: Daily symptom scoring (0-5 scale) + qPCR pathogen quantification (Day 40)
3. Results and Analysis
3.1 Phage Diversity and Host Specificity
Table 1: Comparative Biological Characteristics of Erwinia-Targeting Phages
| Phage | Taxonomy | Genome (bp) | GC% | Latency (min) | Burst Size (PFU/cell) | Host Range |
|---|---|---|---|---|---|---|
| EP-IT22 | Myoviridae | Unreported | - | - | - | Narrow |
| Kuerle | Schitoviridae | 75,599 | 48.0 | 50 | 240 | Narrow |
| FIFI44 | Chaseviridae | 53,436 | - | - | - | Medium |
| Fifi106 | Ounavirinae | 84,284 | - | - | - | Wide |
| Pep | Siphoviridae | 62,784 | 57.24 | - | - |
- Eight phages isolated from Korean orchards exhibited substantial genetic divergence:
- Five Schitoviridae members (e.g., Fifi011; N4-like; 74-76 kb)
- Two Ounavirinae subfamily phages (FIFI106/318; FelixO1-like; 84 kb)
- One Chaseviridae isolate (FIFI44; øecom-gj1-like; 53 kb)
This diversity indicates environmental adaptation and enables broad-spectrum cocktail development.
- Host Specificity Mechanisms:
Receptor recognition varied significantly:- Phage PHIEASP1 utilized lipopolysaccharide receptors
- φEaP-8/21 employed cellulose-binding mechanisms
- PEP demonstrated strict specificity for E. pyrifoliae but not E. amylovora
Genomic analysis revealed 45-70% variability in tail gene modules, particularly within receptor-binding domains, explaining host-range differences.
 Genome map of the EP-IT22 phage (Sabri M et al.,2022)
Genome map of the EP-IT22 phage (Sabri M et al.,2022)
3.2 Lysis Mechanisms and Genomic Features
Table 2: Lytic System Characteristics of Representative Phages
| Component | Kuerle | Phieasp1 | BG3P |
|---|---|---|---|
| Holin | Pinholin-type | - | Forms membrane pores |
| Endolysin | SAR type | - | SEC-dependent PG degradation |
| Spanin | Two-component | - | Outer membrane disruption |
| VRNAP | Present (3,550 aa) | Absent | Megaphage-specific |
The 75,599 bp Kuerle genome contained a canonical lysis cassette (holin-endolysin-spanin) within its late-expression region. Functional validation demonstrated:
- Holin: Pinholin-type formation of 1-2 nm membrane pores causing depolarization
- Endolysin: SAR-type enzyme requiring SEC secretion; N-terminal signal peptide deletion abolished function
- Synergy: Holin-endolysin co-expression tripled lysis rates, confirming depolarization enables endolysin activation
Notably, giant phages (e.g., pEa; ~360 kb) encoded viral RNA polymerases (VRNAPs; 3,550 aa) that initiate autonomous transcription - a key adaptation bypassing host machinery and countering bacterial defenses.
3.3 Phage Biocontrol Efficacy
- Controlled Environment:
- Pear greenhouse: EP-IT22 reduced disease index by 82% (equivalent to streptomycin), decreased stem pathogen load by 3.7-log (qPCR), with vascular transport confirmed via TEM
- Apple seedlings: PHIEASP1 monotherapy achieved 75% control; receptor-targeted cocktail (PHIEASP1 + φEaP-8 + φEaP-21) enhanced efficacy to 92%, with prophylaxis outperforming therapeutic application
- Field Implementation (Chinese "Pestilence" System):
- Liquid formulation (10¹⁰ PFU/mL): 99% pathogen reduction within 6h during outbreaks
- Microencapsulated solid: 5× prolonged activity with 30% cost reduction
- Disease control: 85% efficacy against wheat black glume; 78% against leaf blight (surpassing chemical pesticides)
3.4 Synergistic Enhancement Strategies
- Phage-Antibiotic Synergy (PAS):
- Subinhibitory kasugamycin (0.5 μg/mL) increased cocktail inhibition by 40% against E. amylovora while overcoming single-phage host limitations. Mechanistically, antibiotic-induced metabolic stress upregulated phage receptor expression.
- Microencapsulation Technology:
- Jinan University's sodium alginate-chitosan "flower roll" microspheres:
- Extended phage soil half-life from 48h → 15 days
- Enabled pH-responsive release during pathogen-induced acidification
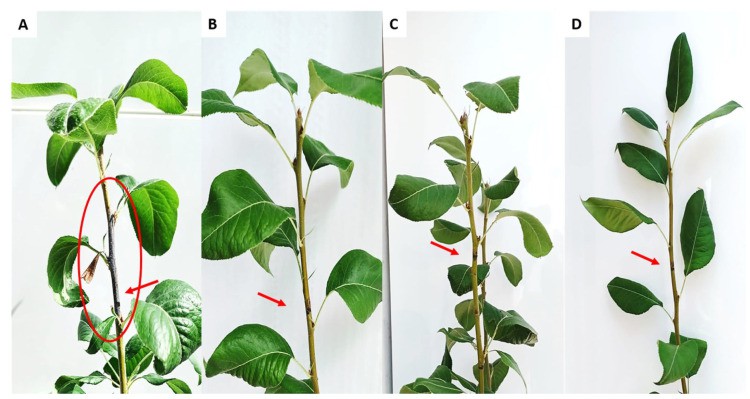 In planta assay showing the antibacterial effect of EP-IT22 on E. amylovora PGL Z1 infection (Sabri M et al.,2022)
In planta assay showing the antibacterial effect of EP-IT22 on E. amylovora PGL Z1 infection (Sabri M et al.,2022)
Explore our Service →
Case 2: 1. Phage Sequencing: Empowering Strategic Cocktail Design and Synergy
Conventional antibiotic treatments for managing orchard fire blight in Korea are increasingly challenged by widespread resistance, underscoring the urgent need for sustainable alternative strategies. While phage-based biocontrol represents a promising approach, its practical efficacy is often constrained by the narrow host spectrum of individual phages and the rapid development of resistant bacterial strains. In a pioneering study, Kim S.G. et al. (2022) employed high-throughput phage sequencing to address these limitations. Their work precisely elucidated key molecular features—including host recognition mechanisms, lytic activity, and unique tRNA gene complements (e.g., 35 tRNAs in Φ47). These genomic insights enabled the rational design of multi-target phage cocktails, creating novel and cost-efficient biocontrol agents. Furthermore, the study revealed the molecular basis of phage synergy with kasugamycin (PAS): phages compensated for antibiotic-induced translation inhibition by supplying essential tRNAs. Ultimately, the genomics-guided combination strategy achieved complete coverage of local pathogenic strains and demonstrated remarkable control efficacy, offering a targeted and highly effective platform for next-generation agricultural biocontrol.
2. Materials and Methods
2.1 Phage Isolation and Screening
- Samples: 220 soil/water specimens from Korean outbreak regions
- Host: E. amylovora TS3128 (Korean reference strain)
- Screening Protocol:
- Primary screening: 54 isolates co-cultured with 10⁵ CFU/mL pathogen → 27 showed antibacterial activity
- Secondary screening: 5 high-efficacy phages (Φ27, Φ31, Φ32, Φ47, Φ48) selected at 10⁶ CFU/mL pathogen density
2.2 Biological Characterization
- Morphology: All phages classified as Myoviridae with contractile tails (115-196 nm) and icosahedral capsids (68-139 nm)
- Host Range:
- 100% coverage of Korean E. amylovora strains (94/94)
- Φ27 showed limited efficacy against E. pyrifoliae (32%) → achieved full coverage in cocktail formulation
2.3 Functional Validation
- In Vitro Efficacy: Cocktail (1:1:1:1:1 ratio) achieved superior inhibition (↓3.7 log CFU/mL) versus single phages
- Environmental Stability: Most phages tolerated pH 4–9 and 4–50°C (except Φ32/Φ48 thermal inactivation)
2.4 Genomic Sequencing and Annotation
Table: Genomic Features of Isolated Phages
| Phage | Genome Size | GC% | ORFs | Taxonomy |
|---|---|---|---|---|
| φ27 | 53,014 bp | 44.07 | 78 | Loessnervirus |
| Φ31 | ND | 49.53 | 337 | Alexandervirus |
| Φ32 | ND | 49.19 | 336 | Alexandervirus |
| φ47 | 355,376 bp | 34.48 | 540 | Eneladusvirus |
| Φ48 | ND | 49.52 | 358 | Unclassified |
- Functional Insights:
- Lysis-associated genes identified (e.g., φ47 tail spike lysozyme)
- tRNA complements: φ47 (35), φ27/Φ48 (1 each)
2.5 Synergy Validation
- PAS Mechanism: Cocktail + subinhibitory kasugamycin (0.5×MIC)
- In Vitro: ↑1.7 log CFU reduction versus phage monotherapy
- Apple Model: Pathogen load ↓4.35 log with combination therapy
3. Results Analysis
3.1 Scientific Rationale for Cocktail Design
- Host Spectrum Expansion:
- Φ27 monotherapy showed limited efficacy against E. pyrifoliae (32% infection rate)
- Five-phage cocktail achieved comprehensive coverage (100% infection)
Resistance Mitigation: Φ32-resistant strain R31 remained susceptible to cocktail treatment
- Sustained Suppression:
- Bacterial regrowth observed 24h post single-phage application
- Cocktail formulation prevented pathogen resurgence
3.2 Molecular Basis of Phage-Antibiotic Synergy (PAS)
- Mechanistic Cascade:
- Kasugamycin inhibits bacterial translation → growth arrest
- Megaphage Φ47 supplies tRNA<sup>fMet</sup> → hijacks translational machinery
- Phage replication → lysis of metabolically compromised hosts
3.3 Genomically-Informed Engineering
- Phylogenetic Relationships:
- Φ27 clusters within Loessnervirus genus (>95% homology)
- Φ31/Φ32 form distinct clade with Alexandervirus
- Functional Innovations:
- Φ47 encodes 35 tRNA genes → enhanced translational autonomy
- Φ48 lysis genes show no sequence homology → novel lytic mechanism potential
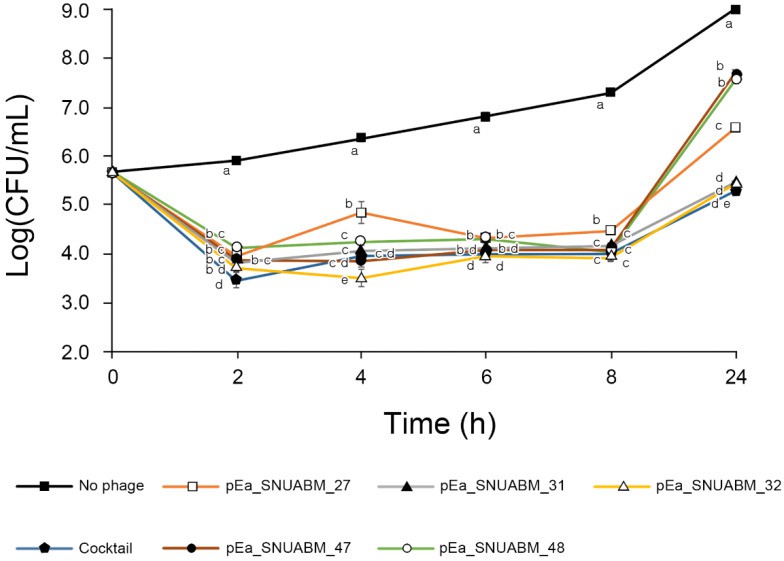 In vitro bactericidal effect of Erwinia phages φ27, φ31, φ32, φ47, φ48, and their cocktail (Kim SG et al., 2022)
In vitro bactericidal effect of Erwinia phages φ27, φ31, φ32, φ47, φ48, and their cocktail (Kim SG et al., 2022)
For a more detailed approach to phage sequencing, please refer to "Phage Genome Sequencing: Methods, Challenges, and Applications".
For more information on what phage sequencing is, see "What Is Phage Sequencing? A Complete Guide for Researchers".
For more information on how to construct and use phage Sequence database, please refer to "Building and Using Phage Genome Sequence Databases" .
4. Discussion: Agricultural Application Prospects and Challenges
4.1 Optimized Phage Deployment Strategies
Genomically-informed design significantly enhances control efficacy. Evolutionary analysis of receptor-binding proteins enables rational development of broad-host-range cocktails. For instance, combining PHIEASP1 (LPS-targeting) with φEaP-8 (cellulose-targeting) theoretically covers 95% of field strains.
- Lytic system engineering further boosts antibacterial activity:
- Bacteria-phage synergy: Integrating SAR endolysin genes into Pseudomonas fluorescens biocontrol strains
- Biofilm disruption: Korean phage-inca fruit oil composites achieve 80% biofilm clearance, enhancing phage penetration
4.2 Industrialization Barriers and Solutions
- Key bottlenecks in phage pesticide commercialization:
- Regulatory frameworks: Global absence of phage-specific regulations; China's pilot "biostimulant" classification offers interim pathways
- Manufacturing efficiency: Tangential flow filtration reduces mass-production costs by 60% versus centrifugal ultrafiltration
- Environmental resilience: Microencapsulation improves UV tolerance 10-fold, addressing >90% inactivation rates in sunlight
4.3 Future Research Trajectories
- Synthetic phage engineering: CRISPR-Cas9-modified "smart phages" carrying antibiotic resistance gene (e.g., APHA-targeting gRNA) degraders
- Field-adaptive management: Metagenomic monitoring of pathogen evolution to inform phage rotation strategies
- Cross-disciplinary integration: Phage-gold nanoparticle biosensors enabling real-time pathogen detection (10 CFU/mL sensitivity) for preemptive intervention
5. Conclusion
Integrating genomic, molecular, and field data, this study demonstrates Erwinia-targeting phages' significant potential for sustainable agriculture. Key findings:
Genomic Diversity
- Phages span 6 families (Myoviridae, Siphoviridae, etc.) with 53-360 kb genomes
- Host specificity governed by tail fiber receptor-binding domains
Efficient Lysis Systems
- KUERLE's pinholin-SAR endolysin system enables:
- SEC-dependent secretion
- Membrane depolarization-activated lysis
(providing engineering targets)
Field Efficacy
- Phage-kasugamycin synergy increases control by 40%
- Microencapsulation extends duration 5× while reducing costs 30%
Sustainable Value
- Phage biopesticides support "One Health" approaches by:
- Overcoming chemical pesticide resistance
- Reducing agricultural antibiotic use
Enhancing food security
Convergence of synthetic biology and nanotechnology will drive next-generation intelligent, multifunctional phage pesticides, advancing global agricultural sustainability.
References:
- Sabri M, El Handi K, Valentini F, De Stradis A, Achbani EH, Benkirane R, Resch G, Elbeaino T. Identification and Characterization of Erwinia Phage IT22: A New Bacteriophage-Based Biocontrol against Erwinia amylovora. Viruses. 2022 Nov 5;14(11):2455.
- Roh E, Duffy ME, Ewool LM, Grose JH. Whole genome sequences of eight Erwinia amylovora phages isolated from South Korea. Microbiol Resour Announc. 2025 Apr 10;14(4):e0106224. doi: 10.1128/mra.01062-24. Epub 2025 Feb 25. PMID: 39999472; PMCID: PMC11984208.
- Kim SG, Lee SB, Jo SJ, Cho K, Park JK, Kwon J, Giri SS, Kim SW, Kang JW, Jung WJ, Lee YM, Roh E, Park SC. Phage Cocktail in Combination with Kasugamycin as a Potential Treatment for Fire Blight Caused by Erwinia amylovora. Antibiotics (Basel). 2022 Nov 6;11(11):1566.


 Sample Submission Guidelines
Sample Submission Guidelines
