Primer-free, multi-locus eDNA surveys from water, soil, and sediments—fast species detection and robust phylogenomics.
Key Takeaways
- GS/MGS are shallow-shotgun strategies that target high-copy loci (cpDNA, mtDNA, rDNA) without primers—well suited to eDNA biodiversity surveys.
- They deliver higher taxonomic resolution and broader clade coverage than single-marker metabarcoding in many groups.
- They shorten time-to-insight for museum/preserved specimens, new field collections, and ancient eDNA (aDNA).
- Clear upgrade paths exist to deep shotgun or long-read when functional genes or strain-level resolution are required.
Introduction
Small samples can yield big ecological pictures. Genome skimming (GS) and metagenome skimming (MGS) recover high-copy genomic regions directly from environmental DNA—without PCR primers—to answer "who is present?" and "how are they related?" across microbes, plants, and animals. By capturing chloroplast, mitochondrial, and ribosomal sequences in the same run, GS/MGS provides a pragmatic balance between scope, resolution, and cost for biodiversity mapping and phylogenetic context in research settings. CD Genomics offers end-to-end GS/MGS services—including sampling guidance, shallow-shotgun library prep, cpDNA/mtDNA/rDNA recovery, and taxonomic/phylogenetic reporting—with optional host-DNA depletion and containerized, version-locked pipelines. When your questions extend to functions or strains, we seamlessly escalate to metagenomic shotgun or long-read (PacBio/ONT) sequencing. All services are for Research Use Only (RUO).
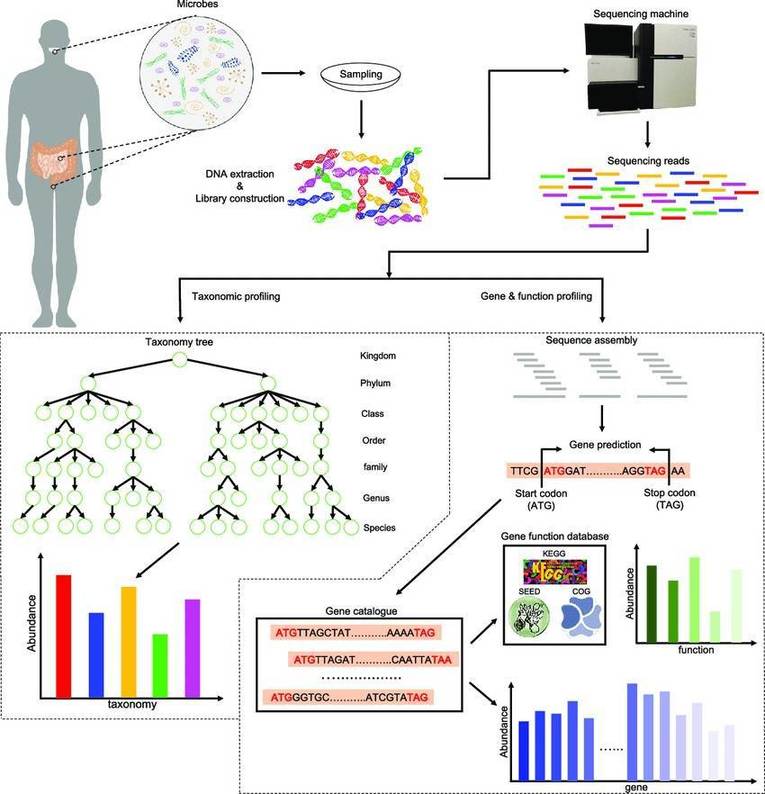
 The workflow of eDNA technology. (Chang, Huihui, et al., Water 2025).
The workflow of eDNA technology. (Chang, Huihui, et al., Water 2025).
What Are GS and MGS?
- Genome Skimming (GS): shallow shotgun sequencing from a single specimen (leaf, tissue, museum voucher) to recover cpDNA, mtDNA, and rDNA for identification and phylogeny.
- Metagenome Skimming (MGS): shallow shotgun sequencing from mixed or environmental DNA (water, soil, sediments) to recover the same loci across taxa within a community.
How they differ from metabarcoding:
Metabarcoding depends on primers and typically targets one barcode (e.g., COI, ITS, 16S). GS/MGS is primer-free and multi-locus, reducing amplification bias, mitigating primer mismatches, and improving taxonomic resolution—often to species or below in groups with good references.
Relationship to deep shotgun:
GS/MGS are optimized for presence and placement. If your questions demand functional genes, metabolic pathways, mobile elements, or strain-level variation, escalate to deep shotgun or long-read metagenomics.
Workflow Overview (Sample → Result)
1. Sampling & Extraction
Environmental matrices (water, soil, sediments) or individual specimens; stabilization strategies and contamination-aware handling.
2. Library Preparation & Shallow Shotgun Sequencing
Short-read libraries sequenced to economical depths tuned to sample complexity and resolution goals.
3. Read Filtering & Host-DNA Management
Optional host depletion (pre- or post-sequencing) to increase effective coverage of target taxa.
4. Mapping/Assembly of High-Copy Loci
Mapping to cpDNA/mtDNA/rDNA references when available; de novo reconstruction for conserved regions when references are incomplete.
5. Taxonomic Identification & Phylogenomics
Multi-locus evidence boosts assignment confidence; extended-barcode trees reconcile signals across loci.
6. Quality Control & Reproducibility
Negative/positive controls, inhibition checks, replicate logic; pipeline versioning and reference release pinning (DOIs, checksums).
Typical outputs: raw reads (FASTQ), locus assemblies/mappings (FASTA/BAM), species lists with confidence metrics, phylogenetic trees, and a methods/QC report detailing parameters and database versions.
Top Applications of GS/MGS in eDNA Biodiversity Research
Museum & Preserved Specimens
GS revives information from degraded or archived material. With appropriate extraction chemistry, fragment-size profiling, and strict contamination control, cpDNA/mtDNA/rDNA can be recovered from silica-dried or ethanol-preserved vouchers to anchor taxonomies and rebuild phylogenies.
What matters most: short-fragment compatibility, completeness across cpDNA/mtDNA/rDNA, and transparent reporting of controls.
 DNA yield against specimen age. (Zeng, CX. et al., Plant Methods 2018).
DNA yield against specimen age. (Zeng, CX. et al., Plant Methods 2018).
Fresh Field Collections (Water, Soil, Sediments)
MGS scales to broad biodiversity audits and can detect divergent taxa that primer-bound assays miss. Handheld or benchtop sequencers support flexible logistics; lab batch-processing enables predictable costs for large campaigns.
Practical pairing: MGS plus full-length 16S/ITS or deep shotgun on a subset to validate key taxa and seed local references.
Ancient Environmental DNA (aDNA)
High-copy loci persist better than single-copy nuclear genes in lake sediments, permafrost, and cave deposits. Expect clean-room practices, damage-pattern checks, multiple negatives, and conservative thresholds. The payoff is deep-time biodiversity snapshots that can be cross-validated with stratigraphy and paleo records.
Extended Barcodes & Phylogenomics
Single-marker barcodes (COI, ITS, 16S) are invaluable but limited by primer bias and locus-specific resolution. GS/MGS builds an extended barcode by combining cpDNA, mtDNA, and rDNA signals:
- Broader detection across distant clades
- Higher resolution among recent divergences
- Robustness against locus-specific dropout
- Greater phylogenetic stability across markers
For lineages with decent reference coverage, extended barcodes can approach deep-shotgun confidence for presence/identity questions at a fraction of the sequencing depth.
Method Comparison: When to Use What
| Dimension | Metabarcoding | GS (single specimen) | MGS (eDNA) | Deep Shotgun |
|---|---|---|---|---|
| Primer dependence | High | None | None | None |
| Loci captured | Single marker | cpDNA, mtDNA, rDNA | cpDNA, mtDNA, rDNA (community) | Genome-wide |
| Taxonomic resolution | Marker-limited | High | High | Highest (incl. functions/strains) |
| Clade coverage | Marker-dependent | Broad | Broadest for eDNA | Broad |
| Degraded DNA tolerance | Medium | High | High | Medium–High (depth-dependent) |
| Reference dependence | Moderate | High (locus refs) | High (locus refs) | High (genome refs) |
| Depth/cost | Low | Low–Moderate | Low–Moderate | High |
| Best use case | Routine biomonitoring | Museum/vouchered IDs | Rapid eDNA audits | Function, pathways, strains |
Rule of thumb: Start with MGS for broad mapping; escalate to deep shotgun or long-read when you need genes, pathways, or strain-level answers.
Challenges and Mitigations
- Reference Database Gaps
Build regional panels; cross-validate with curated global resources; version-lock database releases (DOIs/checksums) for reproducibility. - Depth vs Budget
Tie depth to resolution goals (species vs genus), community complexity, and replicate strategy. Pilot a subset to tune depth before scaling. - Bioinformatics Bias
Benchmark multiple mappers/assemblers; use consensus calls across tools; report identity/coverage thresholds and sensitivity analyses. - Low-Abundance Detection
Increase replicates; optimize extraction; evaluate inhibition (humics, salts) and consider occupancy modeling for rare taxa.
 Challenges of using eDNA in aquatic ecosystems. (Chang, Huihui, et al., Water 2025).
Challenges of using eDNA in aquatic ecosystems. (Chang, Huihui, et al., Water 2025).
Practical Considerations (Bench → Compute)
- Sampling & Preservation: standardize volumes/filters; track blanks; choose stabilization (cooling, ethanol, silica) appropriate to sample type and logistics.
- Host DNA Depletion: enzymatic or capture-based strategies can improve effective coverage—balance benefit vs cost/time.
- Depth Planning: define success by taxonomic confidence and replicate power; shallow depth across more replicates often outperforms deep depth on fewer samples.
- QC & Controls: extraction/library blanks, process controls where appropriate, explicit inhibition testing.
- Reproducibility: containerize pipelines; publish parameter files; pin references with checksums.
- Reporting: describe tools and versions, thresholds, negative-control outcomes, and data availability.
 Future prospects and technological advancements in eDNA. (Chang, Huihui, et al., Water 2025).
Future prospects and technological advancements in eDNA. (Chang, Huihui, et al., Water 2025).
Conclusion
GS/MGS provide a practical, primer-free, multi-locus route to eDNA biodiversity assessment. They excel when the goal is presence and phylogenetic placement across broad clades, and integrate cleanly with deeper approaches when functional inference or strain resolution becomes necessary. With careful sampling, calibrated depth, and transparent QC, GS/MGS can underpin biodiversity baselines, monitoring programs, and hypothesis generation in non-clinical research.
CD Genomics supports GS/MGS studies from design to delivery—depth planning, replicates, library prep, sequencing, and standardized outputs (reads, locus assemblies, species lists, trees, and methods/QC). We can pair GS/MGS screens with deep-shotgun or long-read follow-ups and downstream analyses (e.g., metapangenomics) to match your scientific objectives and budget. Contact us to discuss your samples and goals, or to request a tailored quote and timeline.
For Research Use Only. Not for diagnostic or therapeutic use.
FAQ
- 1) What's the core difference between GS and MGS?
- 2) Why choose GS/MGS over metabarcoding?
- 3) How much sequencing is "enough" for GS/MGS?
- 4) Can GS/MGS handle degraded DNA (museum/aDNA)?
- 5) What if reference databases are incomplete?
- 6) When should I upgrade to deep shotgun or long-read?
- 7) What outputs should I expect?
- 8) How do you control contamination and inhibitors?
References
- Chang, Huihui, et al. "Application of Environmental DNA in Aquatic Ecosystem Monitoring: Opportunities, Challenges and Prospects." Water (20734441) 17.5 (2025).
- Zeng, CX., Hollingsworth, P.M., Yang, J. et al. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods 14, 43 (2018).
- Hoban, M.L., Whitney, J., Collins, A.G. et al. Skimming for barcodes: rapid production of mitochondrial genome and nuclear ribosomal repeat reference markers through shallow shotgun sequencing. PeerJ 10, e13790 (2022).
- Coissac, E., Hollingsworth, P.M., Lavergne, S., Taberlet, P. From barcodes to genomes: extending the concept of DNA barcoding. Molecular Ecology 25, 1423–1428 (2016).
- Dodsworth, S. Genome skimming for next-generation biodiversity analysis. Trends in Plant Science 20, 525–527 (2015).