The integration of molecular markers into plant breeding traces its origins to the late 20th century, when advancements in molecular biology began to unravel the genetic architecture of crops. Early efforts focused on restriction fragment length polymorphisms (RFLPs), which provided the first glimpses into genetic diversity at the DNA level. Over decades, this field has evolved from rudimentary marker systems to high-resolution genomic tools, revolutionizing the precision and efficiency of crop improvement.
In modern plant breeding, molecular markers serve as indispensable tools for accelerating trait selection, mitigating environmental constraints, and addressing global food security challenges. By bridging the gap between genotype and phenotype, they enable breeders to bypass lengthy phenotypic screening processes, particularly for complex traits such as drought tolerance or disease resistance. The advent of marker-assisted selection (MAS) and genomic prediction models underscores their transformative role in developing climate-resilient crops.
Classification of Plant Molecular Markers
Molecular markers have revolutionized plant genetics by enabling precise characterization of genetic architectures across species. This systematic classification explores marker types through their technical principles, historical significance, and emerging innovations in modern breeding programs.
DNA-based molecular markers
Genomic DNA markers remain indispensable tools for deciphering plant genetic blueprints. Early PCR-based techniques like random amplified polymorphic DNA (RAPD) provided rapid polymorphism detection through arbitrary primer amplification, though with limited reproducibility. The development of amplified fragment length polymorphisms (AFLP) enhanced resolution through restriction enzyme digestion and selective nucleotide extensions. Contemporary systems prioritize single nucleotide polymorphisms (SNPs) and simple sequence repeats (SSRs), which combine genome-wide coverage with codominant inheritance patterns. High-density SNP arrays now drive genome-wide association studies in maize, identifying loci controlling drought tolerance through allele frequency shifts across contrasting ecotypes. SSRs continue serving as workhorses in conservation genetics, exemplified by their application in tracing gene flow between wild and cultivated soybean populations across East Asia.
Protein molecular weight markers
Electrophoretic protein profiling laid the foundation for early molecular taxonomy despite technical limitations. Isozyme analysis differentiated plant varieties through migration patterns of catalase or peroxidase variants in native gels, revealing population substructure in perennial species like Quercus. Modern applications extend to validating transgene expression through western blotting, where recombinant proteins are quantified against pre-stained molecular weight standards. Recent refinements in 2D gel electrophoresis coupled with mass spectrometry enable detection of stress-induced protein isoforms in wheat, linking post-translational modifications to salinity adaptation mechanisms.
Codominant vs. dominant markers
Marker inheritance patterns dictate their utility across breeding workflows. Codominant systems like SNPs permit exact zygosity determination, proving critical for studying allelic interactions in hybrid rice heterosis. Breeders leverage this feature to optimize parental line combinations through dosage effect quantification. Conversely, dominant markers such as AFLPs efficiently profile population-level diversity in forest trees, where heterozygosity estimates suffice for conservation prioritization. Emerging multiplex platforms now integrate both paradigms – kompetitive allele-specific PCR (KASP) assays simultaneously genotype hundreds of SNPs while retaining dominant scoring options for high-throughput pre-screening.
Emerging marker systems
Next-generation molecular tools are transcending traditional genetic polymorphism analysis. CRISPR-Cas9-induced insertion-deletion mutations serve as traceable markers in gene-edited crops, enabling non-GMO verification through target site sequencing. Methylation-sensitive AFLP (MS-AFLP) profiles epigenetic variation in vegetatively propagated species like potato, where DNA methylation patterns correlate with tuberization timing under photoperiod changes. Spatial transcriptomics-derived markers now map gene expression gradients in plant tissues, exemplified by identifying auxin biosynthesis hotspots during Arabidopsis root gravitropism. These systems collectively bridge the gap between genetic potential and phenotypic realization in complex breeding environments.
This evolving classification framework reflects the dynamic interplay between technological advancement and biological discovery. From foundational protein variants to epigenome-aware markers, each category addresses unique challenges in plant genome analysis while informing strategic selection in precision breeding initiatives.
Detection Methodologies: Bridging Genotype to Phenotype
The evolution of molecular marker technologies reflects a dynamic interplay between classical methodologies and cutting-edge innovations, each addressing specific challenges in linking genetic variation to observable traits.
Electrophoretic Platforms laid the foundation for early marker systems, relying on gel-based separation to visualize DNA polymorphisms. Techniques like AFLP (Amplified Fragment Length Polymorphism) exemplify this era: by combining restriction enzyme digestion with selective PCR amplification, AFLP generates reproducible fingerprint profiles. While labor-intensive, its independence from prior genomic knowledge makes it indispensable for studying orphan crops such as finger millet, where reference genomes are scarce. Recent adaptations, like capillary electrophoresis, have streamlined AFLP workflows, enabling semi-automated allele scoring in species like cassava for drought tolerance screening.
Complementing these methods, Hybridization Technologies such as Fluorescent in situ Hybridization (FISH) bridge cytogenetics and molecular genetics. FISH employs fluorescently labeled DNA probes to map sequences directly onto chromosomes, offering spatial resolution unattainable through sequencing alone. In wheat breeding, FISH has been instrumental in tracking introgressed segments from wild relatives (Aegilops spp.) into elite lines, enabling precise transfer of rust resistance genes like Sr26. This cytogenomic approach synergizes with next-generation sequencing (NGS), validating structural variations predicted in silico with physical chromosomal evidence.
The advent of SNP Microarray Systems marked a leap toward high-throughput genotyping. Platforms like the Illumina Infinium Wheat 660K SNP array have transformed polyploid crop research, allowing genome-wide association studies (GWAS) in hexaploid wheat to dissect complex traits such as heat-adaptive yield components. These arrays balance cost-efficiency with resolution, making them accessible to public breeding programs. For instance, the IRRI Rice SNP Panel has identified salinity tolerance loci in indica rice varieties, accelerating the development of salt-tolerant cultivars for coastal agroecosystems.
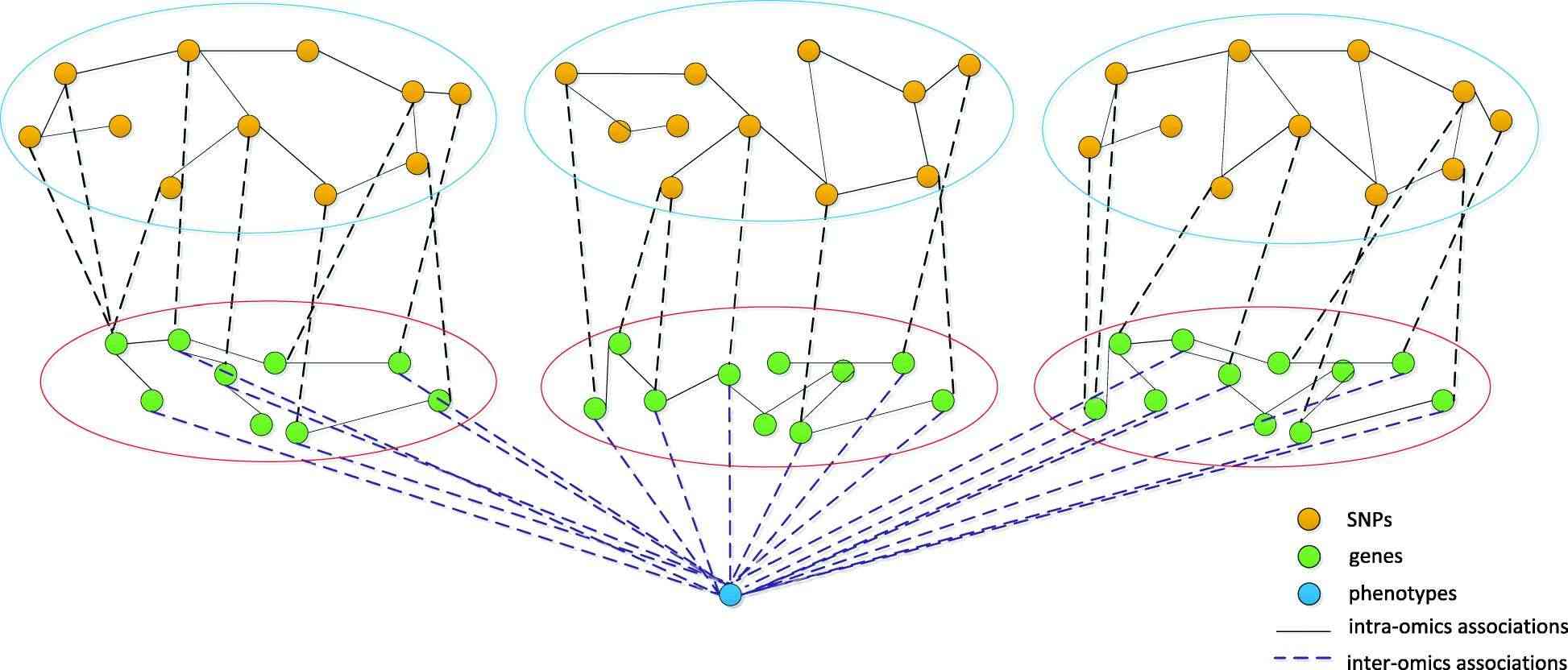
Model of genotype-phenotype association based on data integration algorithm (Guo et al., 2021)
To further enhance scalability, Targeted Sequencing Strategies such as Genotyping-by-Sequencing (GBS) and RAD-seq focus sequencing efforts on reduced genomic regions. GBS, with its multiplexed library preparation, enables cost-effective screening of recombination hotspots in large populations. In maize, GBS-based nested association mapping (NAM) populations have unraveled genetic architecture underlying kernel row number, a key yield determinant. Similarly, RAD-seq has been deployed in sunflower to identify adaptive loci linked to drought escape mechanisms, demonstrating its utility in ecological genomics.
At the frontier of resolution, Single-Molecule Real-Time Sequencing (PacBio) transcends limitations of short-read technologies by capturing full-length transcripts and structural variations (SVs). In maize pan-genome studies, PacBio resolved presence-absence variations (PAVs) affecting flowering time-variants previously obscured by repetitive sequences. These long reads also enable in silico marker development for complex genomes, such as sugarcane, where haplotype phasing is critical for breeding clonal hybrids.
Learn More
Application of Molecular Markers in Plant Breeding
Molecular marker technologies have redefined modern plant breeding by enabling precise genetic manipulation and trait dissection. These tools bridge genomic information with phenotypic expression, offering unprecedented resolution in crop improvement strategies.
Genotype identification and purity assessment
Molecular markers have become a cornerstone for verifying genetic identity in commercial seed production. For hybrid crops like maize, SNP-based assays are routinely employed to confirm hybrid purity by distinguishing parental alleles. In rice, microsatellite markers (SSRs) are used to detect off-types in breeder seed lots, ensuring varietal integrity. A notable example is the use of Kompetitive Allele-Specific PCR (KASP) markers in soybean to authenticate high-value cultivars, reducing economic losses from seed adulteration.
Genetic diversity analysis
By quantifying allelic variation, molecular markers illuminate evolutionary relationships and breeding potential. The 22k SNP array for tomato germplasm revealed cryptic diversity in wild relatives, guiding introgression of heat tolerance traits. Similarly, DArTseq technology has been applied to characterize Ethiopian durum wheat landraces, identifying drought-adaptive alleles absent in modern varieties. These insights inform pre-breeding programs to broaden genetic bases.
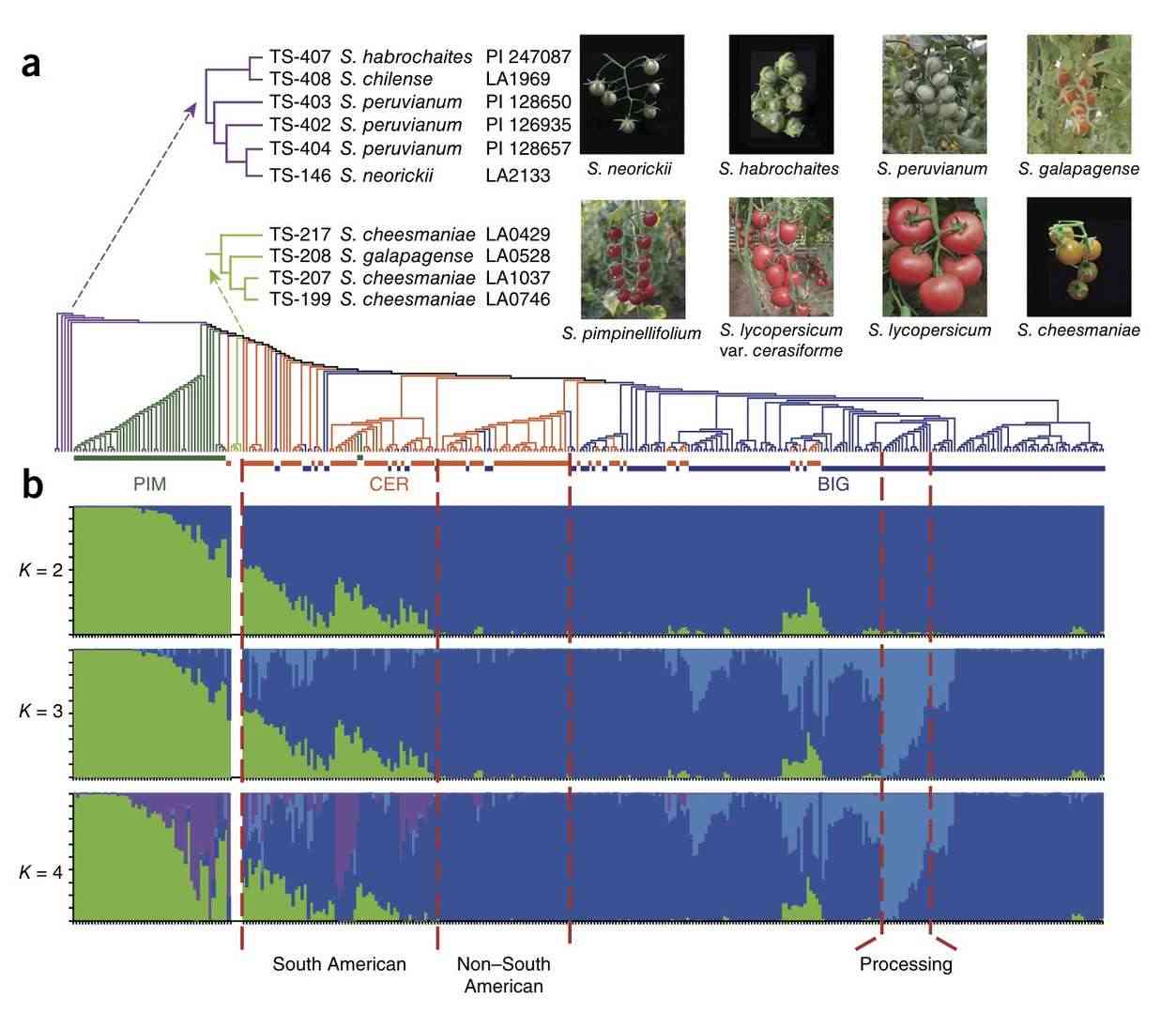
Genome-wide relationship and fruit morphology in cultivated tomato and its wild relatives (Lin et al., 2014)
Marker-assisted selection
MAS accelerates trait introgression by tracking target loci across generations. In wheat, the Sr2 gene marker has been pivotal in pyramiding stem rust resistance genes, while Ppd-D1 allele-specific markers enable photoperiod insensitivity in barley. A breakthrough case involved MAS-driven development of submergence-tolerant rice (Swarna-Sub1), which combines yield stability with flood resilience, benefiting millions of farmers in South Asia.
Genetic map construction and QTL mapping
High-density linkage maps anchor trait-associated regions to chromosomes. The Solanum pennellii introgression lines in tomato facilitated mapping of fruit weight QTLs, leading to the cloning of fw2.2, a key regulator of cell division. Recent pan-genome-based QTL analysis in Brassica napus uncovered structural variants controlling seed glucosinolate content, demonstrating how marker technologies resolve complex metabolic traits.
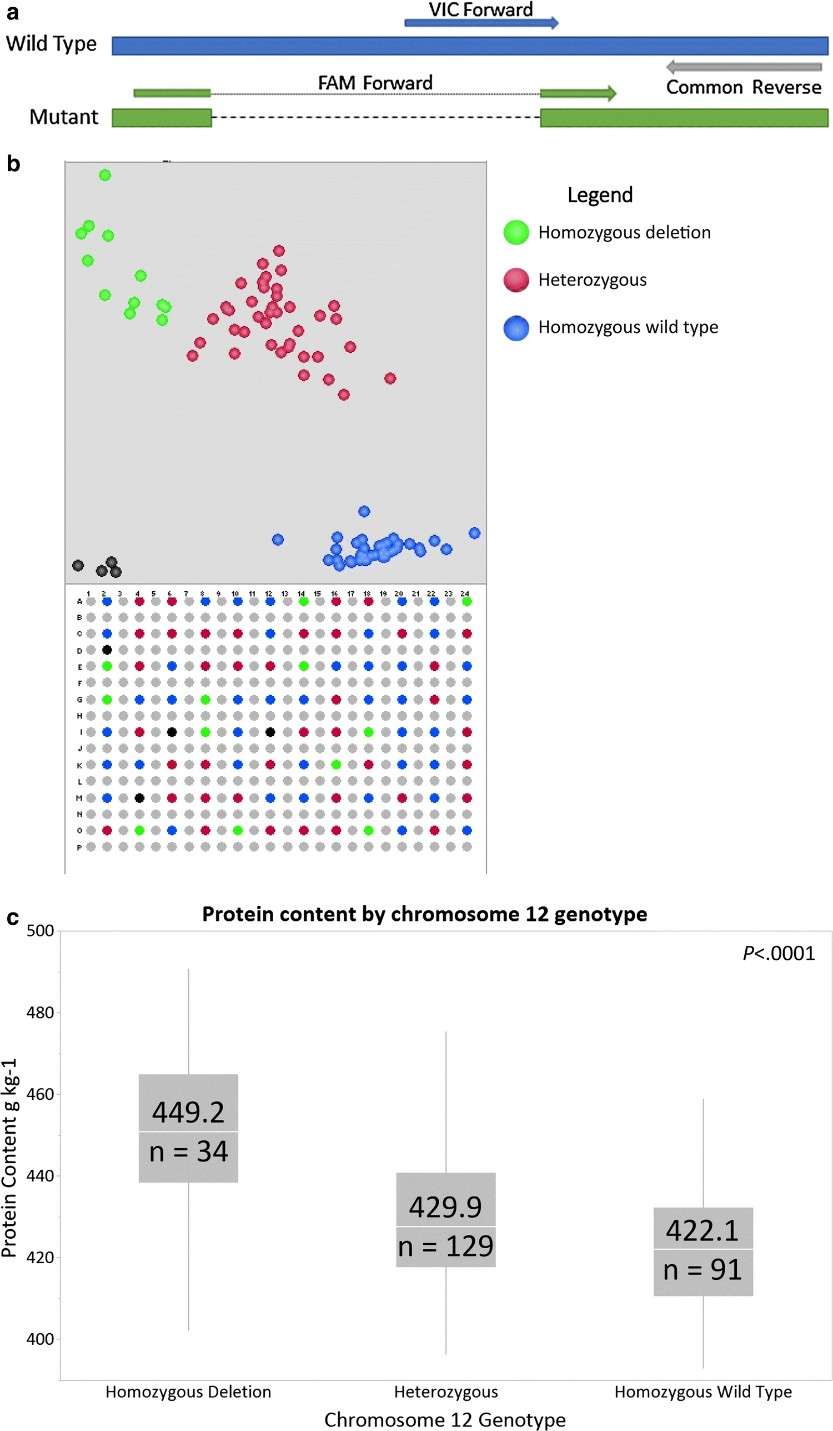
Genotyping of the Benning × G15FN-12 population and associations with seed protein (Prenger et al., 2019)
Challenges and Future Prospects
The harmonization of biological terminology emerges as a foundational pillar for advancing cross-species omics research. Adoption of NIH Gene Ontology (GO) terms, exemplified by resolving ambiguities between broad descriptors like "epigenetic modification" and precise mechanisms such as "DNA methylation," establishes a unified semantic framework essential for interspecies data comparability. This standardization extends to trait-specific nomenclature, where journals mandate systematic naming conventions (e.g., qYLD4.1 for yield-related QTLs), ensuring reproducibility in genetic mapping studies. Such terminological rigor directly supports technical advancements, as seen in mutation breeding programs where space-induced mutagenesis in wheat quantified a 3-fold elevation in SNP frequency over conventional gamma-ray methods, providing empirical benchmarks for optimizing climate-resilient crop development.
Spatially resolved molecular validation further bridges the gap between genomic predictions and phenotypic expression. In cotton fiber studies, the integration of 10x Genomics Visium with single-cell RNA-seq spatially mapped GhEXP1 expression gradients to specific cell wall loosening zones, resolving prior inconsistencies between transcriptomic markers and observed developmental patterns. This multidimensional approach underscores the necessity of combining high-resolution omics layers to decode complex biological processes.
Cross-disciplinary methodologies are redefining scalability in agricultural genomics. Variational autoencoders (VAEs), initially developed for cancer genomics, now enhance genomic selection accuracy in polyploid crops like potato by distilling high-dimensional SNP data into interpretable latent variables. Concurrently, blockchain technology addresses germplasm authentication challenges, as demonstrated by IBM Food Trust’s pilot project tracking heirloom rice varieties from seed banks to commercial markets. By embedding molecular marker data into immutable ledgers, this system safeguards against biopiracy while automating royalty distribution to farmers.
Collectively, these synergistic advances-terminological unification, spatially enhanced omics validation, and computational integration-forge a transformative pathway for precision breeding. They not only address current technical limitations but also establish scalable frameworks for global food security and biodiversity conservation.
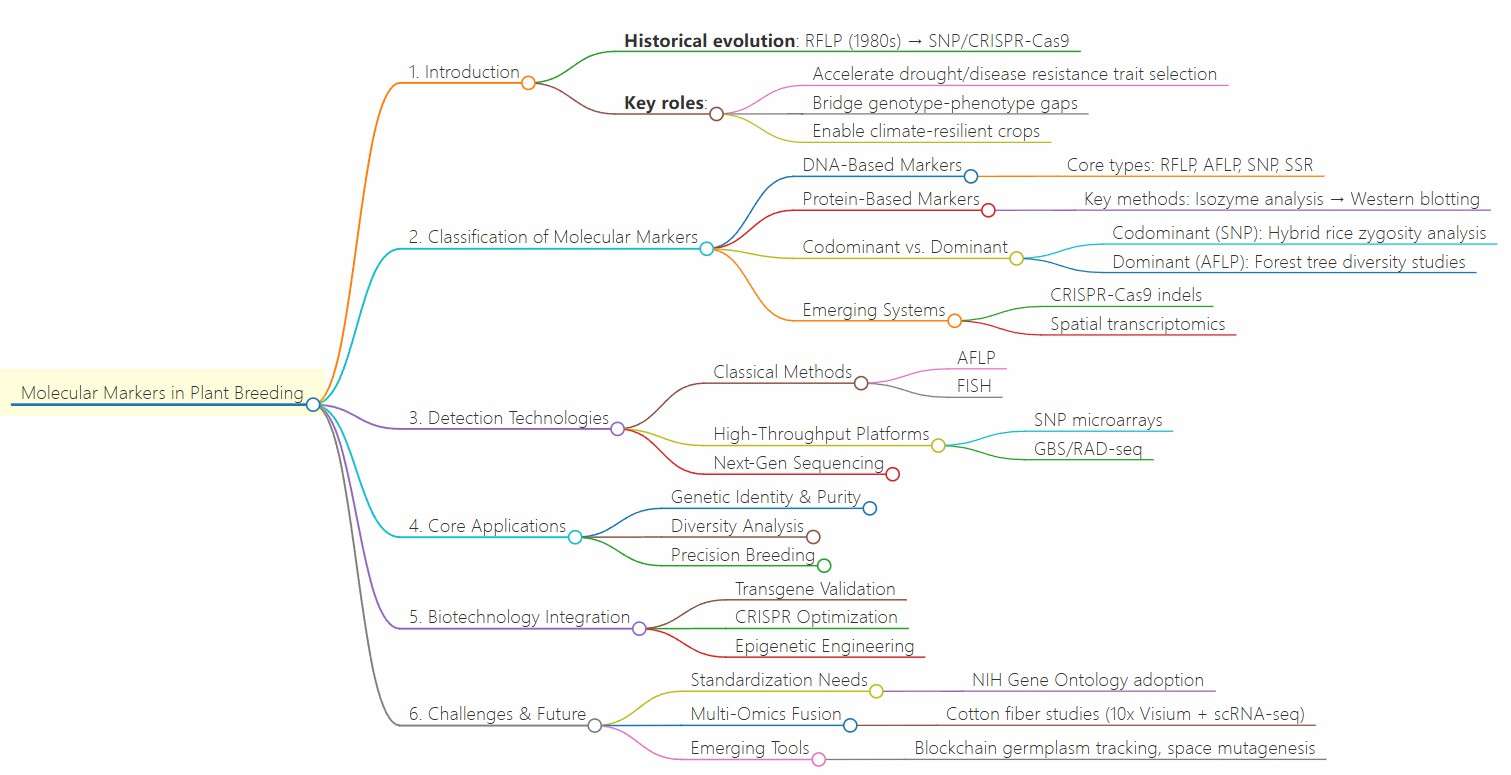
Summary of molecular markers in plant breeding
References
- Guo X, Song Y, Liu S, Gao M, Qi Y, Shang X. "Linking genotype to phenotype in multi-omics data of small sample." BMC Genomics. 2021 22(1):537. https://doi.org/10.1186/s12864-021-07867-w
- Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, Huang Z, Li J, Zhang C, Wang T, Zhang Y, Wang A, Zhang Y, Lin K, Li C, Xiong G, Xue Y, Mazzucato A, Causse M, Fei Z, Giovannoni JJ, Chetelat RT, Zamir D, Städler T, Li J, Ye Z, Du Y, Huang S. "Genomic analyses provide insights into the history of tomato breeding." Nat Genet. 2014 46(11):1220-1226. https://doi.org/10.1038/ng.3117
- Prenger EM, Ostezan A, Mian MAR, Stupar RM, Glenn T, Li Z. "Identification and characterization of a fast-neutron-induced mutant with elevated seed protein content in soybean." Theor Appl Genet. 2019 132(11):2965-2983. https://doi.org/10.1007/s00122-019-03399-w
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. "High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects." Nature. 2016 529(7587):490-495. https://doi.org/10.1038/nature16526


 Sample Submission Guidelines
Sample Submission Guidelines