Comprehensive Overview of GWAS: Definition, Advantages and Methods
Genome Wide Association Studies Definition
Genome-Wide Association Studies, abbreviated as GWAS, represent a scientific methodology aimed at elucidating potential associations between genetic loci, namely the genome, and specific traits or diseases. Through meticulous analysis of millions, even tens of millions, of genetic variations within an organism's genome, GWAS endeavors to identify genetic loci significantly associated with particular phenotypes or diseases. To date, GWAS has successfully unveiled a plethora of genetic variations tightly linked to various phenotypes and diseases, with these findings continuing to accumulate as sample sizes expand and research progresses. The analytical outcomes of GWAS hold broad utility, not only facilitating a deeper understanding of the biological underpinnings of the phenotypes under scrutiny but also enabling assessments of genetic heritability, computations of genetic correlations, clinical risk predictions, and providing crucial research directions for drug development. Furthermore, GWAS aids in uncovering potential causal relationships between environmental factors and specific health outcomes.
The Main Process of GWAS
Data Collection:
DNA samples are systematically collected from individuals, along with recording their phenotypic information, which includes demographic data such as possible disease conditions, age, and gender.
For GWAS, typically large sample sizes are required to identify reproducible genome-wide significant association loci. Determining sample sizes can be facilitated using software tools such as CATS or GPC. The phenotypes involved can be either binary traits or quantitative traits. Additionally, the study design can be population-based or family-based.
Genotyping:
Genotyping of each individual's genes is carried out using existing GWAS arrays or sequencing strategies.
Microarray technology is commonly employed for genotyping common variants, or next-generation sequencing methods such as WES or WGS are utilized to cover rare variants. Given the high cost of WGS (whole genome sequencing), genotyping based on microarrays is currently the most commonly used method. However, WGS theoretically allows for the determination of nearly every genotype in the entire genome, and thus, with the ongoing development of low-cost WGS technologies, it is expected to become the mainstream method in the coming years.
Quality Control:
Rare variants are removed, and variants not meeting Hardy-Weinberg equilibrium tests are filtered out.
SNPs with missing data in a small proportion of individuals in the cohort are filtered out.
Genotyping errors are identified and corrected to ensure consistency between phenotype and genetic data.
Population stratification analysis is conducted.
Phasing and Imputation:
Following the sample and variant quality control of GWAS array data, variants are typically phased, and imputation is performed using a sequenced haplotype reference panel. This involves statistical inference of genotypes that have not been directly analyzed.
Association Statistical Testing:
In GWAS, whether the phenotype is continuous (e.g., height, blood pressure, or body mass index) or binary (e.g., presence or absence of a disease), linear or logistic regression models are commonly used to test the correlation between genotype and phenotype. Covariates such as age, gender, and ancestry are also considered to account for stratification and mitigate confounding effects of population factors.
Meta-Analysis (Optional):
Meta-analysis may be conducted as needed to synthesize results from multiple GWAS.
Result Validation and Interpretation:
Independent replication is sought, and in-depth interpretation and validation of study results are conducted through subsequent analyses post multiple GWAS.
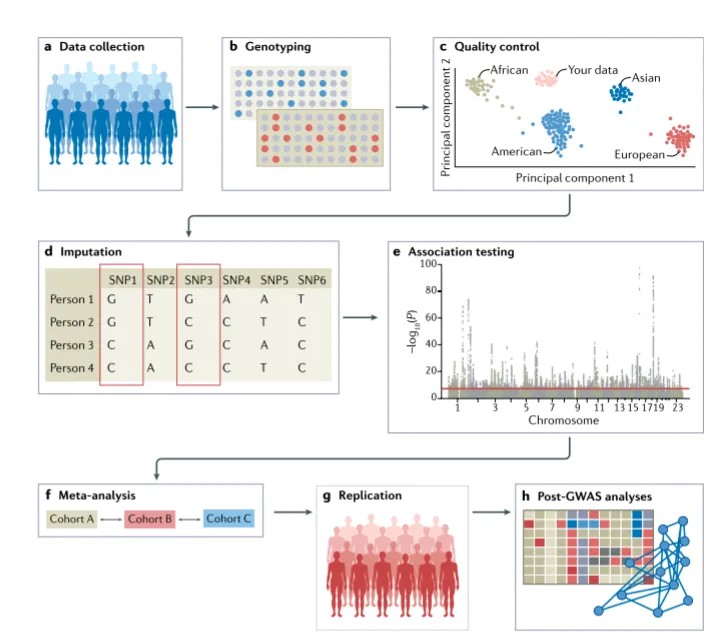 GWAS workflow
GWAS workflow
The Basic Principle of GWAS
GWAS lies a foundational principle that surpasses the confines of conventional genetic inquiry: the methodical exploration of genetic diversity spanning the entirety of the genome. This formidable pursuit is facilitated by the utilization of cutting-edge genotyping methodologies, empowering investigators to concurrently evaluate hundreds of thousands to millions of SNPs dispersed across the human genome. Through scrupulous examination of these genetic loci within expansive cohorts, researchers endeavor to elucidate the nuanced genetic terrain underpinning multifaceted maladies and characteristics.
The Key Advantage of Genome-Wide Association Studies
| Advantage | Description |
| Comprehensive Genome Coverage | GWAS examine millions of genetic variants across the entire genome simultaneously, providing a comprehensive view of genetic architecture. |
| Hypothesis-Free Discovery | GWAS employ a hypothesis-free approach, allowing for unbiased discovery of genetic associations without preconceived hypotheses. |
| High Statistical Power | Large sample sizes in GWAS enhance statistical power, enabling the detection of genetic variants with modest effects on complex traits or diseases. |
| Population Diversity and Generalizability | GWAS investigations across diverse populations ensure the generalizability of findings and uncover genetic factors relevant to different ethnic groups. |
| Replication and Validation | GWAS emphasize rigorous replication and validation of findings, enhancing the reliability and reproducibility of results. |
| Multifactorial Trait Exploration | GWAS enable the investigation of complex traits influenced by multiple genetic and environmental factors, providing insights into the polygenic nature of common diseases. |
| Data Sharing and Collaborative Research | GWAS foster data sharing and collaboration within the scientific community through large-scale consortia and publicly accessible databases, accelerating scientific discoveries. |
You may interested in
Applications of Genome-Wide Association Studies
Unraveling Complex Genetic Architectures
One of the paramount advantages of GWAS lies in their unparalleled ability to unravel the complex genetic architectures underpinning multifactorial diseases and traits. Unlike traditional candidate gene approaches, which often prioritize genes based on prior knowledge or hypotheses, GWAS offers a hypothesis-free, agnostic approach to genetic discovery. By surveying hundreds of thousands to millions of genetic variants scattered across the genome, GWAS can uncover novel genetic loci and pathways implicated in disease etiology, shedding light on previously unexplored biological mechanisms.
Large-Scale Data Acquisition
A cornerstone of GWAS is the acquisition of large-scale datasets comprising thousands to hundreds of thousands of individuals, including both cases (individuals affected by the disease or trait of interest) and controls (unaffected individuals). This vast pool of genomic data provides researchers with unprecedented statistical power to detect subtle genetic associations with diseases and traits. By analyzing such extensive datasets, GWAS can identify genetic variants that confer susceptibility to diseases, thereby facilitating the development of targeted interventions and personalized treatment strategies.
Identifying Common Disease Variants
GWAS excels in identifying common genetic variants that contribute to the risk of complex diseases prevalent in the population. By examining the frequency of genetic variants in cases versus controls, GWAS can pinpoint common alleles that are disproportionately represented in individuals with the disease. This information not only elucidates the genetic basis of common diseases such as diabetes, cardiovascular disorders, and autoimmune conditions but also provides valuable insights into disease mechanisms and potential therapeutic targets.
Accelerating Therapeutic Development
The insights gleaned from GWAS hold immense potential to accelerate therapeutic development and drug discovery. By identifying genetic variants associated with disease risk or treatment response, GWAS can inform the design of targeted therapeutics tailored to specific patient subpopulations. Moreover, GWAS findings can elucidate the underlying biological pathways implicated in disease pathogenesis, providing researchers with valuable targets for drug development and repurposing efforts. In this way, GWAS serves as a vital tool in the quest to translate genetic insights into effective treatments for a wide range of human diseases.
GWAS have become indispensable tools in plant genetics research, offering unparalleled insights into the genetic basis of complex traits in various crop species.
The Power of Genome-Wide Scans
Unlike traditional candidate gene approaches, which focus on specific genes hypothesized to play a role in disease pathogenesis, GWAS adopts a hypothesis-free, agnostic approach to genetic discovery. This comprehensive survey of the entire genome enables scientists to uncover novel genetic loci and pathways implicated in disease etiology, often revealing unexpected relationships and shedding light on previously unexplored mechanisms. Through the meticulous analysis of GWAS data, researchers can unravel the intricate genetic tapestry woven into the fabric of human health and disease.
Leveraging Reference Panels and Imputation
To enhance the scope and resolution of genetic variation interrogation, GWAS leverages reference panels such as the Haplotype Reference Consortium (HRC) and employs advanced imputation techniques. Imputation, a process whereby unmeasured genetic variants are inferred based on patterns of linkage disequilibrium, enables researchers to extrapolate information from genotyped SNPs to unassessed regions of the genome. This expansive imputation pipeline can amplify the number of SNPs tested to nearly 40 million per individual, empowering GWAS to uncover subtle genetic nuances that may have otherwise eluded detection.
Genome-Wide Association Studies in Plants
GWAS has transformed plant genetics research by providing a powerful platform for dissecting the genetic basis of complex traits in crops. From enhancing crop productivity and resilience to uncovering genetic diversity and improving nutritional quality, GWAS offers multifaceted applications with far-reaching implications for agricultural sustainability and food security.
Enhancing Crop Productivity and Resilience
One of the primary applications of GWAS in plants is the identification of genomic regions associated with agronomically important traits, such as yield, disease resistance, and stress tolerance. By analyzing large-scale genotype-phenotype datasets, researchers can pinpoint genetic variants linked to desirable traits, providing valuable targets for crop improvement programs. For example, GWAS has been instrumental in elucidating the genetic basis of yield-related traits in maize, rice, and wheat, leading to the discovery of genes involved in grain size, flowering time, and photosynthetic efficiency.
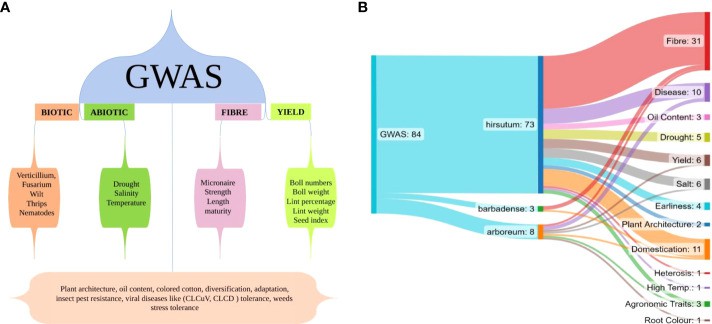 GWAS conducted for different aspects of cotton
GWAS conducted for different aspects of cotton
Accelerating Breeding Programs
GWAS accelerates the breeding process by enabling the selection of superior genotypes based on their genetic makeup. By identifying markers associated with target traits, breeders can efficiently screen germplasm collections to identify elite varieties with desirable traits. This targeted approach to germplasm selection reduces the time and resources required for traditional breeding methods, accelerating the development of new crop varieties with improved agronomic performance.
Uncovering Genetic Diversity and Adaptation
GWAS offers insights into the genetic diversity and adaptation of plant populations to diverse environmental conditions. By analyzing natural variation in crop species, researchers can identify genetic variants associated with adaptation to specific climatic regions, soil types, and biotic stresses. This knowledge enhances our understanding of plant evolution and provides valuable resources for breeding climate-resilient crop varieties. For instance, GWAS studies in rice have revealed genetic loci associated with tolerance to drought, salinity, and heat stress, guiding the development of stress-tolerant rice cultivars tailored to different agroecological zones.
Improving Nutritional Quality and Health Benefits
In addition to agronomic traits, GWAS can elucidate the genetic basis of nutritional traits in crops, leading to the development of biofortified varieties with enhanced nutritional quality. By identifying genes involved in the biosynthesis and accumulation of essential nutrients, such as vitamins, minerals, and antioxidants, GWAS enables breeders to develop crops with improved nutritional profiles. For example, GWAS studies in maize and wheat have identified genetic loci associated with higher levels of micronutrients, such as iron, zinc, and vitamin A, in grain, addressing malnutrition and improving public health outcomes.
Facilitating Precision Agriculture
GWAS contributes to the advancement of precision agriculture by enabling the development of tailored management practices based on genetic information. By identifying genetic variants associated with traits related to nutrient use efficiency, disease susceptibility, and abiotic stress tolerance, GWAS provides insights into genotype-environment interactions, allowing farmers to optimize input use and minimize environmental impact. CD GENOMICS offers comprehensive genomics services to support precision agriculture initiatives, empowering farmers with actionable insights for sustainable crop production.
GWAS vs. Whole Genome Sequencing
By comparing GWAS and WGS across various aspects, researchers can gain a comprehensive understanding of the strengths, limitations, and applications of each approach in genetic research and precision medicine.
| Aspect | GWAS | WGS |
| Scope | Examines genetic variants across the genome, typically using SNP arrays targeting specific genomic regions. | Provides a comprehensive view of the entire genome, capturing all genetic variants including SNPs, insertions, deletions, and structural variants. |
| Resolution | Offers moderate resolution, identifying associations at the level of genetic markers (e.g., SNPs) and genomic regions. | Offers high resolution, mapping genetic variants at the base-pair level, enabling precise localization of disease-associated loci. |
| Variant Detection | Detects common genetic variants present on genotyping arrays, with limited coverage of rare and structural variants. | Detects all types of genetic variants, including rare and structural variants, providing a more complete catalog of genomic diversity. |
| Discovery Potential | Has the potential to uncover associations between known genetic markers and traits of interest, facilitating the identification of common variants with modest effects. | Offers the opportunity to discover novel genetic variants and regulatory elements that may influence complex traits and diseases, expanding the scope of genetic discovery. |
| Population Studies | Often used in large-scale population studies to identify genetic risk factors for common diseases and traits, leveraging the power of large sample sizes and standardized genotyping arrays. | Enables population-wide surveys of genetic variation, offering insights into population structure, ancestry, and genetic diversity, with the flexibility to explore rare variants and population-specific alleles. |
| Precision Medicine | Provides valuable insights into the genetic basis of disease susceptibility and drug response, informing precision medicine initiatives and personalized treatment strategies. | Holds promise for tailoring medical interventions to individual genetic profiles, identifying rare variants with large effects on disease risk or drug response, and guiding personalized treatment regimens. |
| Integration Potential | Can be integrated with other genomic and functional data to enhance interpretation and validation of association findings, leveraging complementary approaches such as transcriptomics, epigenomics, and functional assays. | Offers opportunities for integration with GWAS data to combine the broad coverage of GWAS with the high resolution of WGS, facilitating fine-mapping and functional annotation of disease-associated loci. |
| Cost and Scalability | Relatively cost-effective and scalable for large-scale studies, particularly when using standardized genotyping arrays and leveraging existing datasets and infrastructure. | Can be cost-prohibitive for large-scale studies, particularly when sequencing large cohorts at high depth, requiring substantial computational resources and bioinformatics expertise. |
GWAS analysis tools
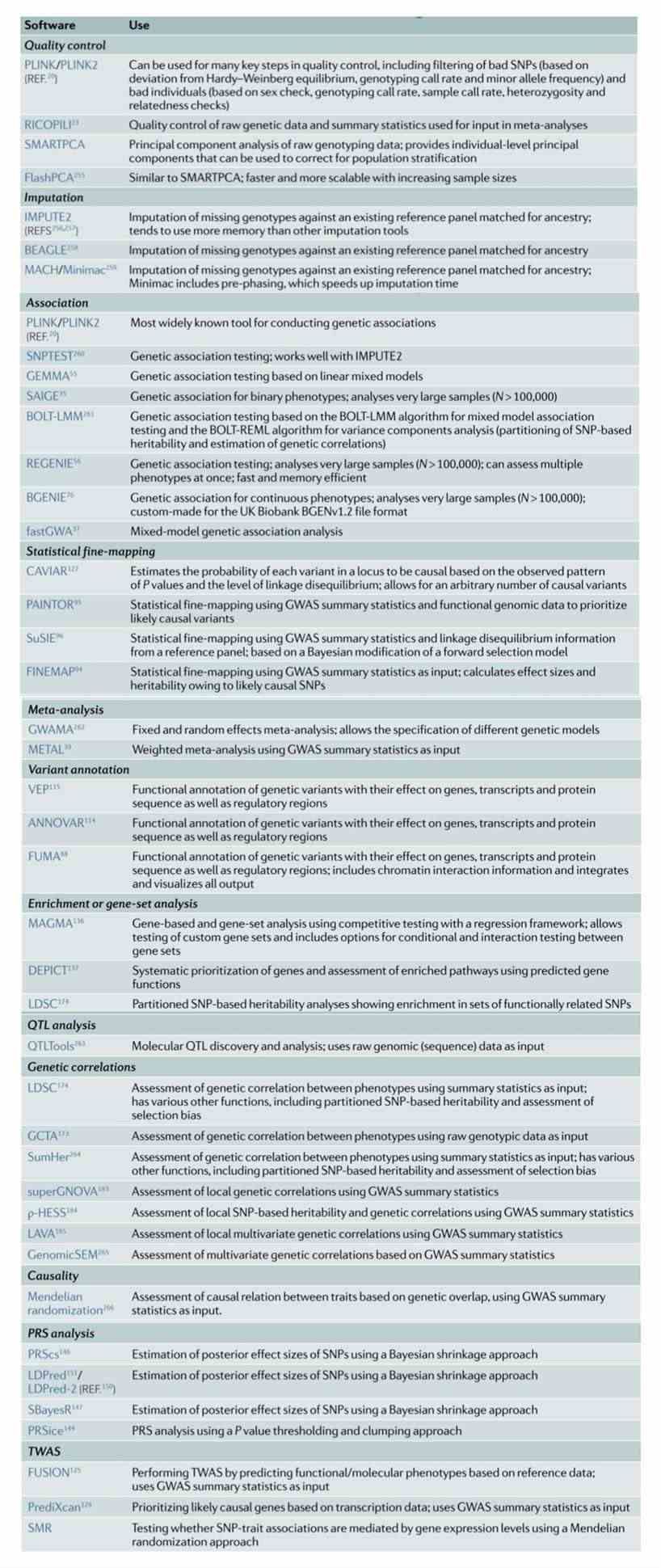
GWAS software analysis tools are integral components in unraveling the intricate genetic foundations of diverse traits and diseases. These sophisticated utilities encompass a wide spectrum of functionalities crucial for executing comprehensive GWAS endeavors. From data preprocessing to association analysis and result visualization, these software suites streamline the intricate process of analyzing extensive genetic datasets.
Researchers heavily rely on acclaimed tools such as PLINK, SNPTEST, and GCTA for their robust capacity to handle genotype data, execute statistical tests, and evaluate genetic associations. Complementing these, advanced algorithms and methodologies provided by tools like EPACTS, SAIGE, and BOLT-LMM enhance the accuracy and efficiency in detecting genetic variants associated with specific phenotypes.
Moreover, software packages such as RVTESTS, EMMAX, and TASSEL offer versatile features for conducting association studies across heterogeneous populations while accounting for various confounding factors. By leveraging these tools, researchers can delve into the genetic architecture of traits and diseases across diverse ethnicities and populations, thereby augmenting our comprehension of genetic diversity and susceptibility to diseases.
Table 1. Commonly used GWAS software analysis tools
What is GWAS database?
A GWAS database functions as a reservoir housing genetic and phenotypic data procured from GWAS endeavors. These repositories meticulously archive information concerning genetic variants dispersed across the entire genome and their correlative patterns with specific traits or maladies.
Central to GWAS databases is the inclusion of genotype data, encompassing details on genetic variations such as SNPs, insertions, deletions, and copy number variations. Concurrently, these databases incorporate phenotypic data, delineating observable traits or attributes of individuals, ranging from disease predisposition to physiological metrics and demographic particulars.
Primarily, researchers leverage GWAS databases to scrutinize the interplay between genetic variants and targeted traits or diseases. Through the interrogation of expansive datasets spanning diverse populations, researchers endeavor to pinpoint genetic determinants implicated in multifaceted traits and diseases, identify putative biomarkers, and unravel the intrinsic biological frameworks underpinning these phenomena.
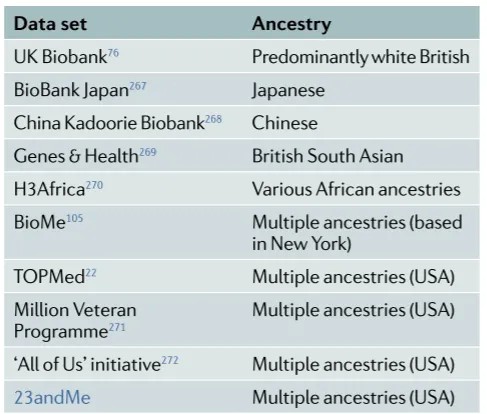
Prominent examples of GWAS databases encompass the Database of Genotypes and Phenotypes (dbGaP), the UK Biobank, the International HapMap Project, the Exome Aggregation Consortium (ExAC), and the 1000 Genomes Project. These repositories serve as invaluable reservoirs for investigators delving into the realms of genetics, genomics, and human health, fostering seamless data dissemination, collaborative ventures, and scientific breakthroughs.
Table 2. Biological Databases with Publicly Available Genotypic and Phenotypic Data
References:
- Uffelmann, E., Huang, Q.Q., Munung, N.S. et al. Genome-wide association studies. Nat Rev Methods Primers 1, 59 (2021).
- Brachi, B., Morris, G.P. & Borevitz, J.O. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol 12, 232 (2011).
- Yasir M, Kanwal HH, Hussain Q, Riaz MW, Sajjad M, Rong J, Jiang Y. Status and prospects of genome-wide association studies in cotton. Front Plant Sci. 2022
- Dash, G.K. et al. (2021). Status and Prospectives of Genome-Wide Association Studies in Plants. In: Gupta, M.K., Behera, L. (eds) Bioinformatics in Rice Research. Springer
- Sánchez-Roncancio C, García B, Gallardo-Hidalgo J, Yáñez JM. GWAS on Imputed Whole-Genome Sequence Variants Reveal Genes Associated with Resistance to Piscirickettsia salmonis in Rainbow Trout (Oncorhynchus mykiss). Genes (Basel). 2022
- Witte JS. Genome-wide association studies and beyond. Annu Rev Public Health. 2010


 Sample Submission Guidelines
Sample Submission Guidelines
