An Overview of SNP Genotyping Technologies
A single nucleotide polymorphism (SNP) represents a variation in a single nucleotide that occurs at a specific position in the genome. SNPs are the major contributors to the DNA variations among individuals, at a frequency of approximately one in 1,000 bp. SNPs have been suggested to be responsible for phenotypic differences, and to affect the development and progression of multiple diseases, as well as to determine the response of drug treatment and/or environmental stress. Additionally, SNPs can serve as ideal molecular markers for identifying genes associated with important biological characters and diseases. Therefore, SNP profiling is considered of great importance in selective breeding, agricultural production and productivity, personalized medicine and drug treatment.
What is SNP genotyping?
SNP genotyping refers to the determination of SNP loci on a whole-genome scale or within genomic regions of interest. The major applications of SNP genotyping are in the treatment of disease and pharmacogenomics studies. A large number of technologies and commercial platforms have been developed for SNP genotyping. According to research purpose, SNP genotyping can be divided into two categories: whole genome association (WGA) and fine mapping. The common approaches for SNP genotyping include SNP microarrays, TaqMan SNP genotyping, MassARRAY SNP genotyping, and high-throughput sequencing. With the rapid development of SNP genotyping technologies, a large number of public databases have been established to collect identified SNPs (Table 1).
Table 1. SNP databases.
| Database | Website |
| dbSNP | http://www.ncbi.nlm.nih.gov/projects/SNP |
| Ensembl | http://www.ensembl.org/Homo_sapiens/index.html |
| Santa Cruz | http://genome.ucsc.edu/cgi-bin/hgGateway |
| SeattleSNPs | http://pga.gs.washington.edu/ |
| PharmGKB | http://www.pharmgkb.org |
| ExPASy Molecular Biology Server | https://www.expasy.org/ |
| EMBL-EBI | http://www2.ebi.ac.uk/ |
| GeneSNPs | http://www.genome.utah.edu/genesnps/ |
| NIEHS SNPs | http://egp.gs.washington.edu/ |
| rSNPBase 3.1 | http://rsnp3.psych.ac.cn/ |
The commercially available arrays for SNP genotyping primarily include Affymetrix SNP and Illumina platforms. The principles of both platforms are outlined in Figure 1. Affymetrix and Illumina genotyping solutions provide trusted performance with respect to precision medicine, agricultural genotyping, forensics, population-scale studies, and genome-wide association studies (GWAS). Commercially available high-density SNP chips cover most of the human genome and other plants and animals of economic significance. Alternatively, semi-custom and custom chips allow researchers to choose desired markers.
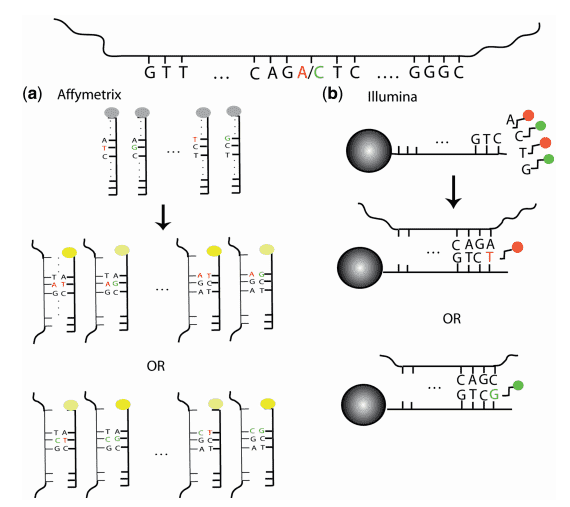 Figure 1. Overview of SNP array technology (laframboise 2009). (a) In the Affymetrix assays, there are probes for both alleles and the DNA binds to both probes regardless of the allele it carries. The impeded binding manifests itself in a dimmer signal. (b) In Illumina beadarray assays, a bead contains a sequence complementary to the sequence adjacent to the SNP loci. When it binds the DNA and exhibits different colors.
Figure 1. Overview of SNP array technology (laframboise 2009). (a) In the Affymetrix assays, there are probes for both alleles and the DNA binds to both probes regardless of the allele it carries. The impeded binding manifests itself in a dimmer signal. (b) In Illumina beadarray assays, a bead contains a sequence complementary to the sequence adjacent to the SNP loci. When it binds the DNA and exhibits different colors.
2. TaqMan SNP genotyping
TaqMan SNP genotyping provides a fast and simple way to analyze SNP sites. TaqMan SNP genotyping assays now comprise more than 17 million pre-designed collections for use in 48-, 96-, 384-, and OpenArray real-time PCR instrument formats. Each pre-designed collection includes two allele-specific TaqMan MGB probes labeled with distinct fluorescent dyes and a PCR primer pair. They uniquely align with the DNA to offer high specificity for the allele of interest. A 5’ reporter dye and a 3’ quencher dye are covalently linked to the probes. When the probes are intact, fluorescence is suppressed. During PCR extension, exonuclease activity happens only on the perfectly matched probes. The reporter and quencher dyes are then released due to the exonuclease activity. The fluorescent signal is measured to determine the genotype of the sample.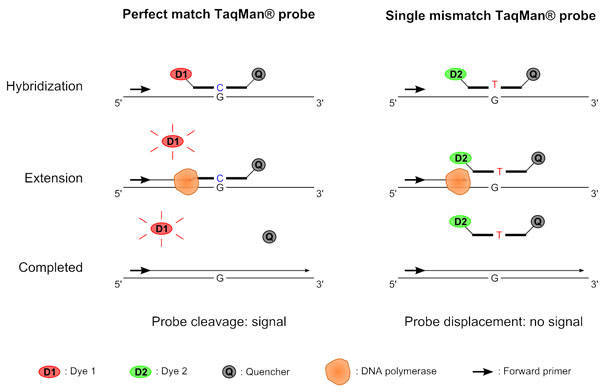 Figure 2. The principle of TaqMan SNP genotyping.
Figure 2. The principle of TaqMan SNP genotyping.
3. MassARRAY SNP genotyping
The MassARRAY platform has several advantages for the users who desire an accurate custom SNP genotyping assay. It utilizes a single extension primer to generate allele-specific products for all PCR sites ensuring a single termination mix. The assay is based on primer extension, and involves a locus-specific primer extension reaction and a locus-specific primer extension reaction (iPLEX assay). In the iPLEX assay, an oligonucleotide primer anneals immediately upstream of the SNP site being genotyped. The primer extension then takes place which is a single complementary mass-modified base. The mass of the extended products is determined by using MALDI-TOF mass spectrometry. MassARRAY SNP genotyping can analysis multiple SNP sites in one reaction. This method is accurate and has a price advantage when analyzing dozens or hundreds of SNPs.
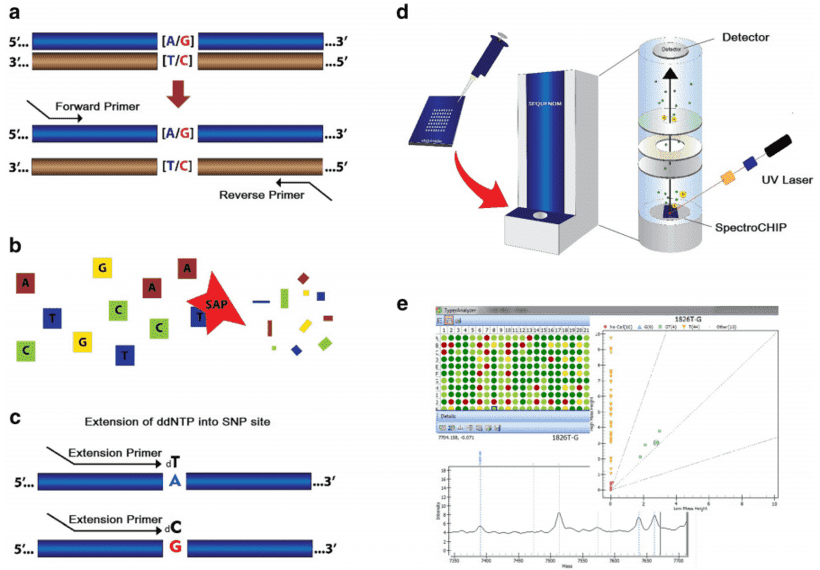 Figure 3. The steps in genotyping using MassARRAY iPLEX system (Svidnicki et al. 2015). (a) Locus-specific amplification reaction. (b) SAP enzyme neutralizes unincorporated dNTPs. (c) iPLEX assay. (d) The products are analyzed by the MALDI-TOF method. (e) Data analysis.
Figure 3. The steps in genotyping using MassARRAY iPLEX system (Svidnicki et al. 2015). (a) Locus-specific amplification reaction. (b) SAP enzyme neutralizes unincorporated dNTPs. (c) iPLEX assay. (d) The products are analyzed by the MALDI-TOF method. (e) Data analysis.
4. SNP analysis by high-throughput sequencing
The sequencing of the human genome identified over 3.1 million validated SNPs. The developed SNP databases provide valuable information for association study design and data interpretation. Sequencing is a powerful and high-throughput approach for SNP genotyping, and a number of methods based on sequencing have been developed for SNP analysis, such as genotyping by sequencing (GBS). SNP analysis by using next generation sequencing(NGS) usually involves the generation of a specific DNA product of selected genomic regions, followed by direct sequencing. With the reduced cost of sequencing, next-generation sequencing (e.g., Illumina, Roche 454, Ion Torrent) and third-generation sequencing (e.g., Nanopore, PacBio SMRT) is becoming widely available.
References:
- Svidnicki M C C C C M, Silva-Costa S M, Ramos P Z, et al. Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iPLEX® technology. BMC medical genetics, 2015, 16(1): 85.
- Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current protocols in human genetics, 2009, 60(1): 2.12. 1-2.12. 18.
- Komar A A. Single nucleotide polymorphisms. Methods in Molecular Biology, 2009, 578.


 Sample Submission Guidelines
Sample Submission Guidelines
