What is a DNA Microarray?
Microarray is a common laboratory tool for detecting gene expression or gene mutations in a high throughput manner. These slides are also known as gene chips or DNA chips. Thousands of probes (with known identity) are immobilized on a microscope slides or silicon chips or nylon membrane, with thousands of tiny spots containing a known DNA sequence or gene. With the advent of DNA sequencing technologies, some tests for which microarrays were used in the past now use sequencing technologies instead. But microarray is less expensive sequencing, so they are still used for very large studies and some clinical tests.
Figure 1. Two type of DNA microarrays.
How does a DNA microarray work?
Depending on the kind of immobilized probes and the information fetched, the microarray experiments can be categorized in three ways: microarrays for gene expression profiling, microarrays for mutation analysis, and microarray-based comparative genomic hybridization.
- Gene expression profiling
To perform gene expression profiling, mRNA molecules are typically collected from both a reference sample and an experimental sample. For example, the reference sample may be obtained from a healthy individual, and the experimental sample may be obtained from a patient. The two sets of mRNA molecules are then converted into complementary DNA (cDNA), with each sample labeled with a fluorescent probe of a distinct color. For instance, the experimental cDNA may be labeled with a red fluorescent dye, and the reference cDNA may be labeled with a green fluorescent dye. The two samples are then mixed together and allowed to hybridize to the DNA probes on the microarray slide. Following hybridization, the microarray is scanned to determine the expression level of genes printed on the slide. If the expression of a particular gene in the experimental sample is higher than in the reference sample, then the corresponding spot on the slide spears red. In contrast, the spot appears green. If the expression level in two samples are equal, then the spot appears yellow. The data obtained through microarrays can be used to generate gene expression profiles, which simultaneously check thousands of genes for expression changes in response to a particular condition or treatment.
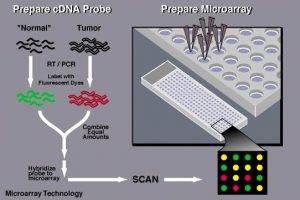
Figure 2. The workflow of DNA microarrays for gene expression analysis.
- Mutation detection
DNA microarray is often used to determine whether a mutation for a particular disease has occurred in an individual. First, researchers obtain a DNA sample from the patient’s blood as well as a control sample without a mutation in the genes of interest. Researchers then denature the DNA in the samples, cut the DNA into small fragments, and label each fragment with a fluorescent dye. While the patient’s DNA is labeled with a green dye, the control DNA is labeled with a red fluorescent dye. Subsequently, the two sets of DNA are inserted into the chip and allowed to bind to the synthetic DNA on the chip. If the individual does not possess a mutation for a gene, both samples will bind to the probes on the chip. If the individual does possess a mutation, the individual’s DNA will not hybridize properly to the probes, while the control sample will bind to the probes on the chip. In addition to the detection of mutations for diseases, DNA microarray can be used for detecting other mutations such as SNP microarray.
- Comparative genomic hybridization
Comparative genomic hybridization (CGH) is used to analyze the entire genome for DNA content different by comparing an experimental sample to a control. The workflow of microarray-based comparative genomic hybridization is similar to microarrays for mutation analysis. But it uses either large-insert genomic clones (BACs/PACs) or PCR products as targets for the hybridization. The primary advantage of microarray-based CGH is the ability to simultaneously identify aneuploidies, deletions, duplications, and amplifications of any locus printed on an array.
Applications of Microarrays
DNA microarrays can be used for many purposes, including gene expression profiling, comparative genomic hybridization, gene ID, chromatin immunoprecipitation on Chip, SNP detection, alternative splicing detection, etc. DNA microarray has been applied to a wide range of fields, such as disease diagnosis, immunogenomics, epigenomics, pharmacogenomics, agricultural genomics, and bioengineering.
References:
- Yu W, Ballif B C, Kashork C D, et al. Development of a comparative genomic hybridization microarray and demonstration of its utility with 25 well-characterized 1p36 deletions. Human Molecular Genetics, 2003, 12(17): 2145-2152.
- Theisen A. Microarray-based comparative genomic hybridization (aCGH). Nature Education, 2008, 1(1): 45.
Further readings:


 Sample Submission Guidelines
Sample Submission Guidelines