What Is Exosome RNA Sequencing and Why It Matters in Modern Biology
1 Introduction — Why Exosomal RNA Is Stealing the Spotlight
Early detection research
In a multicenter pancreatic-cancer study, scientists sequenced small RNAs from blood-borne exosomes and uncovered a transcriptomic signature that flagged stage I–II tumors with an area-under-the-curve of 0.93—using nothing more than a routine blood draw .
Therapy-response profiling
A separate team tracking gastric-cancer patients on PD-1 blockade found that just two exosome-derived miRNAs (miR-451a and miR-142-5p) stratified likely responders vs non-responders in a pilot cohort, pointing to a minimally invasive way to study immune modulation in real time(DOI: https://doi.org/10.21037/tcr-24-2151) .
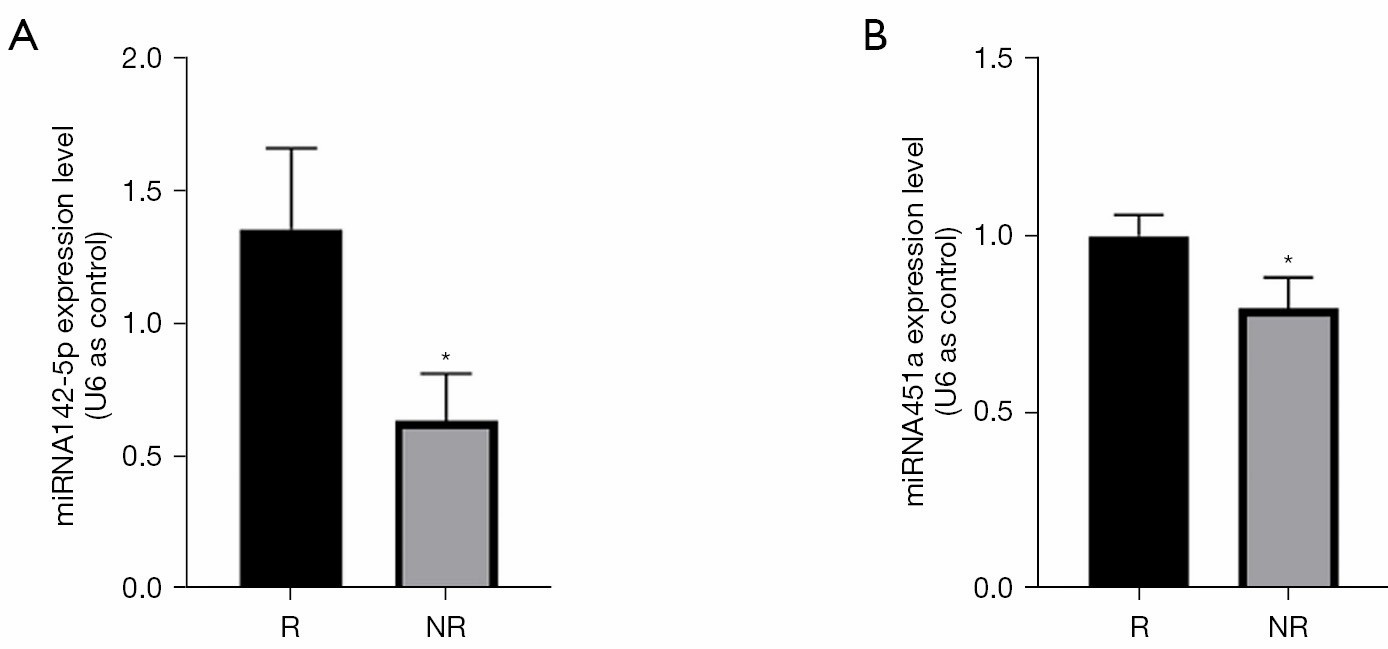 MiRNA451a and miRNA142-5p were upregulated in the responder group.
MiRNA451a and miRNA142-5p were upregulated in the responder group.
These examples highlight a broader shift: researchers no longer have to carve out tissue to follow disease biology. Whereas conventional biopsies offer only a static, local snapshot—and carry obvious procedural burdens—liquid biopsies tap circulating analytes that mirror system-wide events and can be sampled repeatedly with minimal risk (Haozhou Tang et al,. 2024). Among the liquid-biopsy options, exosomes stand out because their lipid bilayer protects RNA cargo from degradation and reflects active cell-to-cell communication rather than passive cell death.
The punchline
All of this momentum converges on a single enabling method: Exosomal RNA Sequencing (exosome RNA-Seq). By capturing the complete RNA payload—mRNAs, lncRNAs, miRNAs, and more—exosome RNA-Seq lets researchers map signaling networks, uncover low-abundance transcripts, and monitor molecular changes over time. In the sections that follow, we'll unpack what exosomes are, how the sequencing workflow works, and why this approach is rapidly becoming indispensable across oncology, immunology, and beyond.
2 Exosomes and Their RNA Cargo
What they are
Exosomes are 40-160 nm, lipid-bilayer vesicles that bud from the endosomal multivesicular-body pathway and are released by virtually every cell type into biofluids such as blood, urine, saliva, and cell-culture medium (Kalluri & LeBleu, 2020. DOI: https://doi.org/10.1126/science.aau6977). Their protected membrane makes the molecular freight—nucleic acids, proteins, lipids—remarkably stable outside the cell, enabling "liquid snapshot" sampling without invasive tissue collection.
Why RNA matters
Inside each exosome is a selectively packaged library of messenger RNAs (mRNAs), long non-coding RNAs (lncRNAs), microRNAs (miRNAs), circular RNAs, and other small transcripts. Because cargo loading is an active, regulated process, the RNA profile mirrors the physiological state and signaling intent of the donor cell rather than random cellular debris.
Functional proof
The field pivoted in 2007 when Valadi and colleagues showed that exosomal mRNAs and miRNAs can be delivered to recipient cells and translated, establishing exosomes as bona-fide vehicles of genetic exchange (Valadi et al., 2007. DOI: https://doi.org/10.1038/ncb1596) . Subsequent work has mapped hundreds of exosome-borne miRNAs that modulate pathways ranging from immune checkpoint regulation to neuronal differentiation.
Research utility
For researchers, this means that exosomal RNA provides a non-invasive readout of cell–cell communication and disease-related signaling. For example, a 2024 systematic review catalogued exosomal miRNA panels that correlate with tumor stage, treatment response, and mechanistic pathways in multiple cancer models—highlighting their growing role as discovery-phase biomarkers rather than clinical diagnostics (Zhu et al., 2024. DOI: 10.1097/MD.0000000000040082) .
3 How Exosomal RNA Sequencing Works
Below is the end-to-end workflow most labs (including ours) follow. Each step has multiple validated options, so you can tailor the protocol to sample type, RNA species of interest, and instrument availability.
| Step | Key Options & Best Practices |
|---|---|
| 1. Sample collection & initial processing | Start with plasma, serum, urine, saliva, or conditioned medium. Keep samples on ice and add RNase inhibitors; freeze at –80 °C within 2 h to preserve vesicle integrity. |
| 2. Exosome isolation |
|
| 3. RNA extraction | Phenol/TRIzol protocols work, but silica- or silicon-carbide-membrane kits (e.g., exoRNeasy or SeraMir) improve small-RNA retention and remove inhibitors. A next-generation ATPS workflow combined with TRIzol recently out-performed legacy methods for picogram-level inputs.( https://doi.org/10.3892/ijmm.2017.3080 ) |
| 4. Library construction |
|
| 5. Sequencing platforms |
|
| 6. Bioinformatics & interpretation | QC → trim → align (STAR/Bowtie2 for mRNA; miRDeep2 or miRge3.0 for miRNA) → count & normalize (DESeq2/edgeR) → functional analysis (GSEA, KEGG, GO). Results are cross-checked against EV-specific databases such as ExoCarta and Vesiclepedia. |
Why this workflow excels
Sensitivity. Vesicle-protected transcripts remain intact, allowing reliable detection of RNAs expressed at <10 copies/cell.
Specificity. Affinity-based isolation plus strand-specific library prep preserves biologically relevant signal.
Scalability. Kit-based extraction and dual-indexed libraries enable high-throughput studies without sacrificing data quality.
Our Exosomal RNA-Seq service delivers everything from vesicle isolation through pathway-level reporting—so you can focus on the biology, not the benchwork.
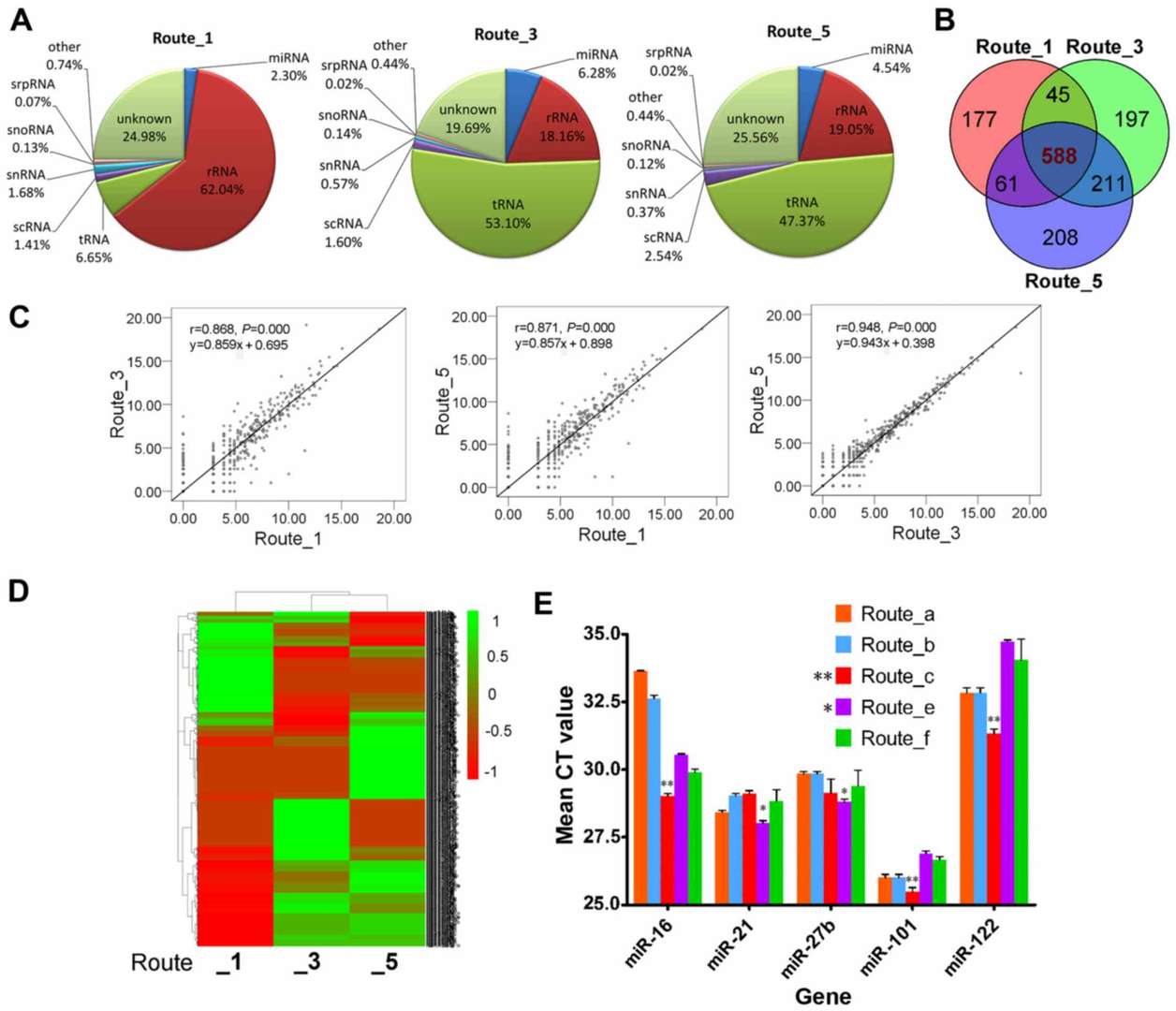 Analysis of the differences in exosome miRNA profiles among the different isolation methods(https://doi.org/10.3892/ijmm.2017.3080)
Analysis of the differences in exosome miRNA profiles among the different isolation methods(https://doi.org/10.3892/ijmm.2017.3080)
4 Exosomal RNA-Seq vs. Other RNA Profiling Strategies
Selecting the right sequencing strategy depends on the biological question, sample accessibility, and the resolution you need. The comparison below highlights where exosomal RNA-Seq shines—and where alternative approaches still rule.
| Exosomal RNA-Seq | Circulating-free RNA-Seq (cfRNA-Seq) | Conventional Tissue RNA-Seq | |
|---|---|---|---|
| Sample source | Plasma, serum, urine, saliva, culture medium | Plasma/serum supernatant | Fresh-frozen or FFPE tissue slices |
| Molecular snapshot | Protected vesicle cargo that reflects active inter-cell signals | Fragmented RNAs passively released from dying cells | Local transcriptome of sampled lesion |
| Key strengths | Stable RNA; captures low-abundance regulatory transcripts; ideal for longitudinal, minimally invasive sampling | High concordance with tissue mutations; useful when vesicle isolation is not possible | High depth; retains spatial context if combined with laser-microdissection or spatial-omics |
| Typical applications | Tumor micro-environment studies, drug-response monitoring, neuro-inflammation models | Early cancer screening panels, host-response signatures for infection (e.g., TB AUC 0.95) | Mechanistic pathway mapping, cell-type deconvolution, validation of liquid-biopsy hits |
| Principal challenges | Isolation bias, vesicle heterogeneity, need for dedicated QC metrics | RNA highly fragmented; lower stability; more background noise | Invasive collection; single-time-point view; can miss heterogeneity |
Why exosomal RNA often wins for non-invasive biology
Signal over noise. Because their lipid bilayer shields transcripts from plasma nucleases, exosomal RNAs show higher integrity and reproducibility than naked cfRNA, improving differential-expression power in low-input studies (Zhu et al., 2024. DOI: 10.1097/MD.0000000000040082) .
Functional relevance. Vesicle cargo is selectively loaded, mirroring the donor cell's communication intent, whereas cfRNA largely derives from apoptotic or necrotic debris.
Longitudinal sampling. Serial phlebotomy or urine collection lets researchers track therapy-induced transcriptome shifts without repeat biopsies—crucial for time-course or drug-screen projects.
That said, cfRNA-Seq remains attractive when rapid, vesicle-agnostic screening is sufficient, and tissue RNA-Seq is still unbeatable for depth and spatial context when surgical samples are available.
You may interested in
Learn more
5 Why Exosomal RNA-Seq Matters — Key Advantages & Emerging Trends
Truly non-invasive, yet information-rich
Because exosomes circulate in virtually every biofluid and shield their RNA cargo from nucleases, they deliver intact transcriptomic snapshots without a scalpel. A 2024 hepatocellular-carcinoma study, for example, built a three-gene exosomal RNA panel that distinguished early-stage HCC from hepatitis and healthy controls with an AUC = 0.85 using nothing more than 2 mL of plasma . Such performance is impossible with fragmented circulating-free RNA and would otherwise require tissue biopsy. (DOI: 10.1186/s12885-024-13332-0)
Enables longitudinal, real-time biology
Serial blood or urine draws let researchers follow molecular trajectories across treatment cycles, disease flare–quiet phases, or developmental timelines. Multi-center efforts now profile EV-RNA at multiple time points to predict resistance to checkpoint inhibitors and to refine dosing windows—work that simply cannot be done with destructive tissue sampling .
Scales from single pathway to pan-cancer discovery
When the question is broad—Which transcripts discriminate eight different tumor types?—exosomal RNA-Seq still delivers. A 2025 study across 1,385 participants built a 12-gene exosomal signature (ETR.sig) that classified eight cancers with macro-average AUC = 0.983, underscoring its power for large-cohort, machine-learning pipelines(DOI: https://doi.org/10.1186/s12943-025-02271-4) .
Extends beyond oncology
Exosomal RNA is proving just as valuable in neuroscience and immunology. Single-cell sequencing of an engineered anti-inflammatory exosome (Exo-srIκB) showed targeted dampening of aged-brain microglia and macrophage signaling, illuminating vesicle-mediated control of neuroinflammation . Similar vesicle-omic studies are mapping stroke progression, autoimmune flares, and even plant–microbe interactions.
Ready for multi-omics integration
Because exosome isolation co-purifies lipids and proteins, the same prep feeds proteomics, lipidomics, or metabolomics—making EV-RNA a natural hub for systems biology projects and pathway-level modeling.
Exosomal RNA-Seq combines the accessibility of a liquid biopsy with the depth of a full transcriptome, delivering actionable, time-resolved insights that conventional cfRNA-Seq or tissue RNA-Seq can't match—all strictly for research, discovery, and pre-clinical development.
6 How Our Exosomal RNA-Seq Service Accelerates Your Research
Our end-to-end platform removes the technical friction between a promising hypothesis and publishable data—so your team can spend its time interpreting biology, not troubleshooting protocols.
What we handle for you
- Sample logistics & integrity
- Exosome isolation & QC
- Low-input library construction
- Sequencing, data processing & analytics
Interactive deliverables
- Raw FASTQ, aligned BAM, count matrices, interactive HTML dashboard, publication-ready figures (PNG & SVG)
- Built-in filters to flag contaminants, hemolysis markers, or library-prep artifacts
Why researchers choose us
- Matrix versatility – validated for >10 biofluids and conditioned-medium systems
- Consistent QC – ≥80 % reads map to EV-RNA reference; duplicate rate <5 % in >95 % of libraries
- Transparent turnaround – standard projects ship data in 15 business days from sample receipt
- Research-use-only compliance – no clinical claims; all protocols follow research-grade SOPs


 Sample Submission Guidelines
Sample Submission Guidelines
