How to Prepare Samples for Exosome RNA Sequencing: A Step-by-Step Guide
Introduction: A Successful Exosomal RNA Project Starts with the Right Sample Prep
In exosomal RNA sequencing, the most common point of failure isn't library construction or data analysis—it's the sample.
Despite advances in RNA-Seq technologies, poor sample preparation remains a top reason for failed or suboptimal results. In fact, internal QC audits and published literature suggest that over 50% of sample failures trace back to errors in early-stage handling, including improper collection, degradation during transport, or contamination from cell-free RNA (cfRNA).
Why is exosomal RNA (exoRNA) so vulnerable?
Low abundance: exoRNA is present in nanogram or even picogram quantities.
Small fragment size: most exoRNAs are short—particularly miRNAs, which are easily lost during extraction.
Susceptibility to degradation: delays in processing or improper freezing can irreversibly damage RNA quality.
Sensitivity to contamination: unlike intracellular RNA, exoRNA prep must contend with free-floating RNA, proteins, and lipids in biofluids.
That's why proper sample prep isn't a minor detail—it's the foundation of your entire project. Whether you're planning a biomarker discovery study, mechanistic investigation, or simply validating vesicle-associated transcripts, preparing your samples correctly ensures you'll get reliable data the first time.
This article provides a standardized, step-by-step guide to help you:
- Choose the right sample type for your study
- Avoid common pitfalls in collection and preservation
- Decide whether to isolate exosomes yourself or submit raw biofluids
- Prepare your samples for submission to our sequencing facility
By following these guidelines, you'll reduce delays, improve data quality, and stay on track toward your research goals.
If you're new to exosome workflows, start here: Exosome Isolation to miRNA Extraction Protocol
Step 1: Choose the Right Sample Type
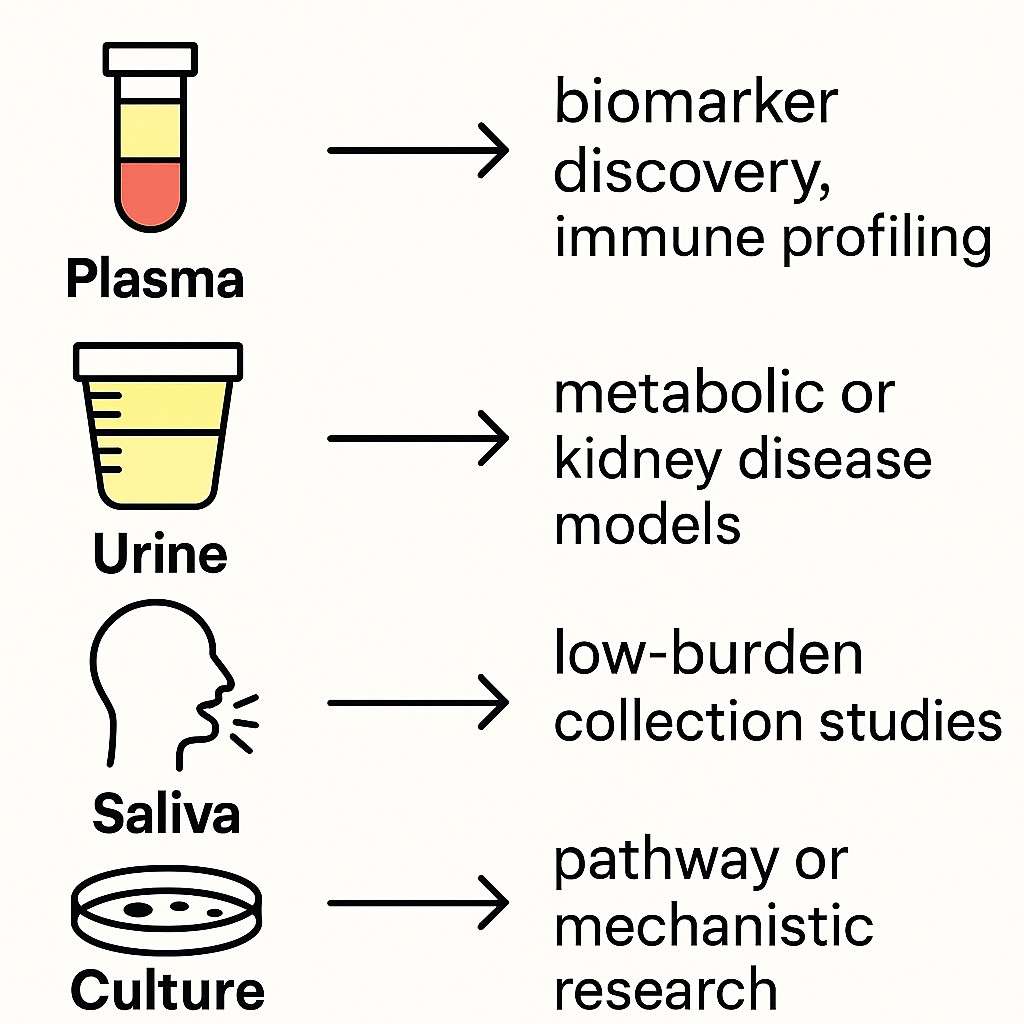
Not all biofluids are equally suited for exosomal RNA analysis. The success of your RNA-Seq project often begins with choosing the sample type that matches your research objectives, input requirements, and exosomal RNA yield expectations.
Below is a comparison of common sample types used in exosomal RNA studies:
| Sample Type | Recommended Level | Use Case Summary |
|---|---|---|
| Plasma (EDTA tube) | ★★★★☆ | High exosome content; widely used in oncology, immunology, and liquid biopsy studies. |
| Urine | ★★★☆☆ | Non-invasive; suitable for nephrology, urology, and metabolic disease research. |
| Saliva | ★★☆☆☆ | Convenient collection; requires rapid processing to prevent degradation. |
| Cell culture supernatant | ★★★★★ | Cleanest vesicle background; ideal for mechanistic studies and drug response screens. |
Plasma (EDTA)
Why it's preferred: Plasma collected with EDTA is a gold standard in exosomal RNA research due to its high vesicle yield and compatibility with both miRNA and mRNA profiling. It's also relatively stable and well-documented across studies.
Best for: Cancer research, immune monitoring, infectious disease models, and general liquid biopsy development.
Avoid heparin tubes—residual heparin inhibits downstream PCR and NGS workflows.
Urine
Why it's useful: Urine is easy to collect non-invasively and is rich in vesicles from the kidney and urinary tract. However, exoRNA content tends to be lower than plasma, and urine is more susceptible to degradation.
Best for: Studies focused on renal function, metabolic syndromes, or non-invasive biomarker exploration.
Saliva
Pros & cons: While easy to collect and patient-friendly, saliva degrades quickly and contains abundant RNases. It also has variable exosome yield across individuals and time points.
Best for: Feasibility studies or niche applications where other fluids are unavailable.
Cell Culture Supernatant
Why it's ideal: Exosomes from culture media are highly pure, with minimal background from cfRNA or plasma proteins. Researchers can control timing, stress conditions, and drug exposures.
Best for: Mechanistic studies, drug testing, pathway analysis, and time-course experiments in vitro.
Choosing the right sample sets the tone for your entire project. In the next section, we'll dive into best practices for sample collection and preservation—a critical step in maintaining exoRNA integrity.
You may interested in
Learn more
Step 2: Sample Collection and Storage Best Practices
Exosomal RNA is highly sensitive to degradation, contamination, and environmental stress. Ensuring proper handling from the moment of sample collection is critical to maintaining RNA integrity and securing high-quality sequencing results.
This section outlines key do's and don'ts for sampling, storage, and transport—based on best practices established in exosome research and validated across hundreds of successful projects.
General Guidelines
Always use RNase-free, sterile consumables for tubes, pipette tips, and filters. RNase contamination is a leading cause of exoRNA degradation.
Label samples clearly with waterproof markers or printed stickers that withstand freezing. Avoid adhesive tape that detaches at −80°C.
For Plasma Samples
Recommended tube: EDTA (lavender top).
Avoid heparin—it interferes with PCR and library prep.
Processing time: Centrifuge within 2 hours of collection to separate plasma from blood cells. Delays can lead to increased cfRNA release from lysed cells.
Aliquot immediately into cryovials (200–500 µL per tube). Do not refreeze thawed plasma.
For Urine, Saliva, and Culture Supernatant
Urine: Centrifuge to remove cells and debris within 1 hour. Add preservatives if delay expected.
Saliva: Use collection kits that include RNase inhibitors, and freeze immediately.
Cell culture media: Collect supernatant at predefined timepoints, centrifuge at 2,000–3,000 g to remove cell debris, and filter if needed.
Storage Recommendations
Temperature: Store all samples at −80°C. Use cryovials rated for ultra-low temperatures.
Aliquoting: Divide into single-use volumes to avoid freeze–thaw cycles.
Maximum hold time: For best results, submit samples within 4–6 weeks of collection if stored properly. Older samples may require QC pre-evaluation.
Shipping and Transport
Preferred method: Ship on dry ice, using foam-insulated containers.
Packaging:
Layer 1: Cryovials inside a sealed plastic bag
Layer 2: Bag placed in a secondary sealed box (e.g., 50 mL conical tubes)
Layer 3: All materials packed in dry ice within a foam-insulated shipper
Include a detailed sample manifest with sample names, volumes, and collection dates.
Following these best practices helps ensure that your samples arrive intact, and that the RNA we extract is suitable for downstream analysis—whether you're targeting small RNAs like miRNAs or full-length transcripts.
Next, we'll explain your two options for submitting samples: raw biofluid or pre-isolated exosomes.
Step 3 – Prepare Your Own Exosomal RNA: What You Must Know Before Submission
CD Genomics specializes in exosomal RNA sequencing services and requires clients to submit already extracted and purified exosomal RNA. We do not provide exosome isolation or RNA extraction services. To ensure the success of your project, please adhere to the following submission requirements.
What You Need to Submit
Sample Type: Purified exosomal RNA, free from proteins, DNA, and enzymatic inhibitors.
Quantity: Minimum of 20 ng per sample.
Concentration: At least 1 ng/µL.
Purity:
OD260/280 ratio between 1.8 and 2.2.
OD260/230 ratio ≥ 2.0.
Integrity: RNA Integrity Number (RIN) ≥ 6.5.
Elution Buffer: RNase-free water or 10 mM Tris-HCl (pH 8.0), without EDTA or other inhibitors.
Storage: Store at -80°C; avoid repeated freeze-thaw cycles.
Reference: CD Genomics Sample Submission Guidelines
What to Avoid
To prevent issues that could compromise your sequencing results:
Do Not Submit:
Whole biofluids (e.g., plasma, urine, saliva, or cell culture supernatant).
Unprocessed exosomes or vesicle pellets.
Avoid:
Elution buffers containing high salt concentrations, SDS, guanidinium, or phenol residues.
Buffers with EDTA, as it can inhibit downstream enzymatic reactions.
Repeated freeze-thaw cycles of RNA samples.If you're uncertain about your buffer composition or extraction method, please contact us for a free compatibility check before submission.
Recommended Isolation & Extraction Practices
While CD Genomics does not endorse specific brands, we recommend that your workflow includes:
Exosome Isolation: Utilize ultracentrifugation, polyethylene glycol (PEG) precipitation, or affinity-based methods.
RNA Extraction: Employ protocols optimized for small RNAs, ensuring minimal carryover of organic solvents.
Quality Control: Assess RNA integrity using a Bioanalyzer or TapeStation to confirm RIN values.
Documentation Required
When submitting your samples, please provide:
A brief summary of your exosome isolation and RNA extraction protocols.
Details of the buffer composition used in the final RNA sample.
Concentration measurements and the methods used.
This information assists us in assessing input compatibility and optimizing your library preparation strategy.
Frequently Asked Questions (FAQ)
What is the minimum amount of RNA required for sequencing?
We recommend a minimum of 20 ng total RNA per sample, at a concentration of ≥1 ng/µL. For small RNA or low-input workflows, please contact us for feasibility review.
Can I send raw biofluid or isolated exosomes?
No. CD Genomics only accepts purified exosomal RNA. Please extract RNA before submission. For recommended extraction guidelines, refer to our Sample Submission Guide.
What buffer should I use to resuspend my RNA?
Use RNase-free water or 10 mM Tris-HCl (pH 8.0). Do not use buffers with EDTA, phenol, guanidinium, or high salt, as they can inhibit downstream enzymes.
Do you offer RNA integrity and quantification services?
Yes. All submitted RNA undergoes QC by Bioanalyzer or TapeStation (RIN ≥6.5 recommended), plus concentration and purity checks via NanoDrop and Qubit. You'll receive a full QC report.
Can you work with degraded or low-input RNA?
We evaluate degraded or low-input samples on a case-by-case basis. If your sample falls below standard thresholds, please contact us in advance for a free technical consultation.
I used a commercial RNA extraction kit—will it work?
In most cases, yes. However, please share your extraction method and buffer details to help us assess compatibility. Some commercial kits leave inhibitory residues that must be avoided.
How should I package and ship RNA samples?
Store RNA at –80°C and ship on dry ice, using leak-proof tubes in a three-layer insulated container. Avoid repeated freeze-thaw cycles.
Conclusion: High-Quality Samples Yield High-Quality Data
In exosomal RNA sequencing, your data quality is only as good as your sample preparation. From choosing the right biofluid to ensuring proper storage and buffer compatibility, every detail matters. Poor handling can lead to low yield, high degradation, or unusable results—wasting valuable time and budget.
Recommended Next Reads
What Is Exosome RNA Sequencing?


 Sample Submission Guidelines
Sample Submission Guidelines
