Overview of Nanopore Sequencing Technology
Abstract
Genes encode the entire hereditary information of an organism, and sequencing is the process of converting DNA chemical signals into digitally computable signals, making sequencing indispensable in genomic research. The advent of Sanger sequencing (first-generation sequencing) propelled the realization of the Human Genome Project, marking significant strides in genomic sequencing technology over the past few decades. The continual advancement of next-generation sequencing (NGS) technologies has led to a progressive reduction in the cost of genomic sequencing, along with increases in sequencing throughput and accuracy, facilitating the analysis of genome structure and the study of genomic genetic variation, thereby greatly advancing genomic research. However, in NGS sequencing, errors introduced by PCR amplification during library construction and the typically short sequencing read lengths (often less than 500bp) frequently impede the technology from meeting the higher demands of some modern biological inquiries. For instance, challenges arise in determining longer repetitive segments on DNA and in assessing DNA/RNA methylation modifications. Consequently, the emergence of third-generation sequencing technologies, such as Nanopore Sequencing, addresses these limitations. Nanopore Sequencing enables the determination of longer read lengths (up to 10kbp) without the need for complex library construction processes. In recent years, Nanopore Sequencing has garnered widespread attention from the scientific community.
Overview of nanopore sequencing
1. What is the nanopore sequencing?
Sequencing primarily involves the identification of the four nucleotide bases in DNA. Due to the striking chemical similarities between purine and pyrimidine bases, distinguishing them poses significant challenges. Currently, the presence of the four bases is primarily converted into signals such as light, electrical, or pH signals, and differences in signal amplification are utilized to identify the different bases. Nanopore sequencing, however, converts the four bases directly into electrical signals, allowing for their identification through variations in current signals and waveform patterns. Moreover, nanopore sequencing differs from traditional first and NGS techniques, which involve concurrent synthesis and sequencing. Nanopore sequencing directly sequences individual molecules, obviating the need for PCR amplification during sequencing, thus effectively mitigating systematic errors resulting from PCR biases. Originating in the 1980s, nanopore sequencing has continually advanced with the development of nanopore proteins and motor proteins, ultimately culminating in its realization.
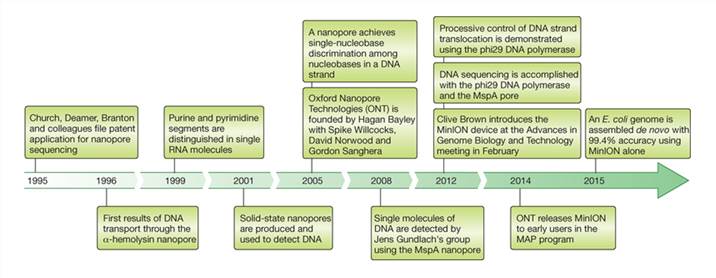 Milestones in nanopore DNA sequencing (David et al.,2016)
Milestones in nanopore DNA sequencing (David et al.,2016)
2. How nanopore sequencing works?
The working principle of nanopore sequencing
In nanopore sequencing, nanopore proteins are immobilized on a polymeric membrane and submerged in an electrolyte solution, with only the nanopores on the membrane allowing ions to pass through. By applying a stable potential difference across the nanopores using an external power source, electrolyte ions pass through the nanopores, generating an electric current across the membrane. Guided by motor proteins, double-stranded DNA (dsDNA) molecules (or RNA-DNA hybrid duplexes) are first unwound, allowing single-stranded DNA or RNA to translocate through the nanopore from the negative (cis) to the positive (trans) side. Due to the differing volumes and charge properties of nucleotides, their passage through the nanopore induces distinct changes in the electric current across the membrane, enabling the identification and sequencing of DNA sequences.
The first nanopore protein discovered to influence ion currents and enable their detection via DNA or RNA was α-Hemolysin, a membrane protein derived from S. aureus. Another commonly used nanopore protein is MspA, with pore diameters close to 1-2 nm. However, in the absence of nanopore proteins, DNA translocation occurs too rapidly to obtain meaningful electric current signals, necessitating the introduction of motor proteins to control the rate of DNA translocation. In 2012, researchers found that phi29 DNA polymerase, when employed as a motor protein in conjunction with the aforementioned nanopore proteins, effectively regulates DNA translocation, yielding clear, high-resolution signals.
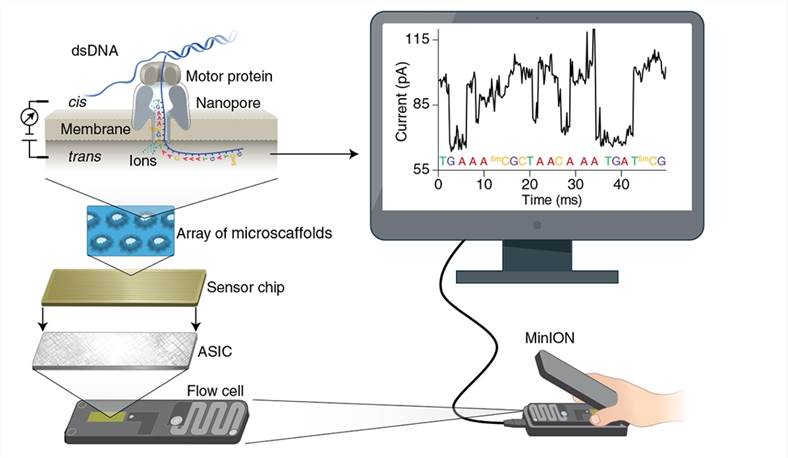 Principle of nanopore sequencing (Wang et al.,2021)
Principle of nanopore sequencing (Wang et al.,2021)
The workflow of nanopore sequencing

DNA extraction: Extraction of DNA or RNA from organisms or tissues, followed by purification.
Library construction: End repair and adapter ligation are performed on DNA or RNA. These adapter sequences contain specific sequences that interact with the nanopore protein on the nanopore sequencing device, allowing molecules to be accurately guided through the nanopore. Subsequently, quality control and fragment size selection are conducted to ensure that the size and quality distribution of molecules in the library meet the requirements.
Load and sequencing: The constructed library is loaded into the nanopore sequencing device for sequencing. Within the device, molecules are guided through the nanopore, and DNA or RNA sequences are sequenced by monitoring changes in electrical current.
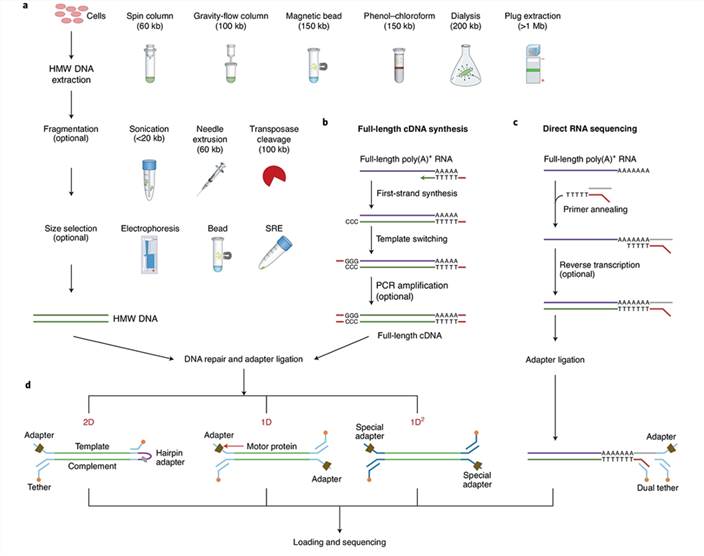 Library preparation workflow for Oxford Nanopore Technologies (ONT) (Wang et al.,2021)
Library preparation workflow for Oxford Nanopore Technologies (ONT) (Wang et al.,2021)
3. Powerful nanopore sequencing applications
The application in genome sequencing
The utilization of nanopore sequencing technology in genome sequencing is extensive. It can be employed to enhance existing reference genomes, with the human reference genome being a prime example. Currently, nanopore sequencing has filled in 12 gaps (>50 kb each) in the human reference genome, including sequences of telomeric repeats and Y chromosome centromeres. Apart from the human genome, the reference genomes of other species can also be expanded and refined using nanopore sequencing. By accurately identifying repetitive regions, for instance, the reference genome of Caenorhabditis elegans has been expanded by over 2 Mb. Nanopore sequencing has also been utilized to read non-reference genomes previously unreported, such as assembling the first genome of Rhizoctonia solani using nanopore sequencing data. For RNA viruses, ONT can also sequence them, such as Zika virus, influenza virus, among others. During the COVID-19 pandemic, ONT aided in swiftly determining the complete viral genome of SARS-CoV-2 through cDNA sequencing and direct RNA sequencing, making significant contributions to subsequent vaccine development efforts.
The application in epigenetics
The role of nanopore sequencing in epigenetic research is increasingly prominent. Independent studies as early as 2013 demonstrated the ability to distinguish methylated cytosines (5mC and 5hmC) in DNA using characteristic current signals in MspA nanopore sequencing. Subsequently, the development of bioinformatics tools enabled the identification of three DNA modifications (6mA, 5mC, and 5hmC) from ONT data. Methods like MeSMLR-seq and SMAC-seq, combining ONT sequencing with exogenous methyltransferase treatment, have been devised to map nucleosome occupancy and chromatin accessibility. Additionally, ONT sequencing allows simultaneous characterization of various epigenetic features on single long human DNA molecules, including endogenous 5mC methylome and nucleosome occupancy. Experimental approaches like DiMeLo-seq and BIND&MODIFY also integrate ONT sequencing with diverse biochemical techniques to map histone modifications, histone variants, and specific protein-DNA interactions.
The application in cancer
Nanopore sequencing is integral to cancer research, spanning various cancer types such as leukemia, breast cancer, brain tumors, colorectal cancer, pancreatic cancer, and lung cancer. Its primary role lies in identifying critical genomic variations, especially those that are intricate and extensive. For instance, in patients with chronic lymphocytic leukemia, nanopore amplicon sequencing effectively detects TP53 mutations. Furthermore, by combining Cas9-assisted target enrichment with nanopore sequencing, detailed analyses of the breast cancer susceptibility gene BRCA1 and its adjacent regions, covering a span of 200 kb, provide insights into the complete range of genetic alterations in relevant disease genes. Nanopore sequencing also directly identifies DNA modifications, capturing genomic and epigenomic changes in brain tumor samples, thereby offering a versatile and rapid molecular diagnostic tool for cancer. Additionally, MinION cDNA sequencing enables the same-day detection of fusion genes in clinical samples. The efficiency of this technology streamlines the entire process, from sample collection to bioinformatics analysis to customized treatment strategies, all achievable within a single day. Beyond cancer, nanopore sequencing holds considerable promise in investigating infectious and hereditary diseases alike.
CD Genomics offers flexible and cost-effective Nanopore sequencing services, tailored to meet your research interests with customized analysis workflows. Each step of the process involves skilled professionals to ensure quality control and result accuracy.
Learn More
4. What are the advantages of nanopore sequencing?
Nanopore sequencing boasts several advantages:
- Sequencing samples are extremely easy to prepare.
- The read length is exceptionally long, reaching several million base pairs, exceeding second-generation sequencing by 1,000 times, and also surpassing third-generation Pac Bio sequencing (tens of thousands of base pairs).
- It can directly sequence RNA molecules and detect epigenetic modifications, such as DNA methylation.
- The cost is low, approximately $150 per experiment (run).
- Sequencing speed is fast, with sample preparation taking approximately 10 minutes and sequencing around 40 minutes.
FAQ
1. What is the difference between Illumina and Nanopore?
Illumina and Nanopore are two widely recognized sequencing methodologies, each characterized by unique principles and operational approaches.
Illumina sequencing, categorized as second-generation sequencing, follows the principle of concurrent synthesis and sequencing. This method entails fragmenting DNA into shorter sequences, followed by parallel sequencing with high throughput. Despite its commendable accuracy and cost-effectiveness, Illumina sequencing typically produces relatively short read lengths, typically ranging from a few hundred to a few thousand base pairs.
In contrast, Nanopore sequencing presents a distinctive approach, directly sequencing DNA or RNA molecules through nanopores, eliminating the need for PCR amplification. During Nanopore sequencing, as DNA passes through minute nanopores, changes in electrical current induced by nucleotides are captured and processed to generate DNA sequences. This technique offers the notable advantage of ultra-long read lengths, extending to millions of base pairs. Furthermore, Nanopore sequencing streamlines the process and facilitates rapid data analysis in real-time. However, it is important to note that Nanopore sequencing may exhibit slightly reduced accuracy compared to Illumina sequencing.
2. What is oxford nanopore sequencing?
Oxford Nanopore Sequencing (ONS) stands as a cutting-edge DNA and RNA sequencing technology pioneered by ONT. At the heart of this technique lies the utilization of a nanoscale protein pore embedded within a highly resistant polymer membrane. During the sequencing process, a constant voltage is applied across the membrane, allowing negatively charged single-stranded DNA or RNA molecules to traverse through the nanopore. As these nucleic acid molecules pass through the nanopore, they induce fluctuations in ion current, which correspond to the nucleotide sequence traversing the nanopore and can be decoded in real-time using computer algorithms, enabling instantaneous DNA or RNA sequencing.Nanopore sequencing technology boasts several notable features including long read length、real-time sequencing、direct RNA sequencing and portability.The application of ONT spans diverse fields including genomics, transcriptomics, epigenetics, and infectious disease research. As the technology continues to advance, its scope and impact are anticipated to expand further.
3. Is nanopore sequencing the most accurate?
Nanopore sequencing technology represents an innovative approach to DNA sequencing, offering distinct benefits that traditional methods often lack. Its notable features include exceptionally long read lengths and the ability to perform real-time data analysis, distinguishing it from conventional sequencing techniques. However, while nanopore sequencing excels in these areas, its precision may not always match that of alternative methods.Regarding accuracy, nanopore sequencing frequently exhibits higher error rates compared to established platforms like Illumina sequencing. Illumina platforms typically produce short read data with error rates ranging from 0.1% to 1%, whereas nanopore sequencing may yield error rates between 6% to 15%. Consequently, applications demanding utmost precision, such as single nucleotide polymorphism (SNP) detection or small-scale variant analysis, may find nanopore sequencing suboptimal.
To mitigate accuracy issues, researchers employ diverse strategies, including multiple sequencing iterations of the same DNA molecule to generate consensus sequences, employing hybrid error-correction approaches that integrate long and short read data, and advancing base-calling algorithms. Despite these efforts, nanopore sequencing's accuracy remains constrained by its single-molecule sequencing nature, characterized by a relatively low signal-to-noise ratio at the individual molecule level.
In summary, nanopore sequencing presents unparalleled advantages in specific applications, particularly those necessitating long read lengths and real-time analytics. Nevertheless, its precision may not surpass that of certain established sequencing technologies. Consequently, the choice of sequencing methodology hinges on the precise requirements and objectives of the research endeavor.
References:
- Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34(5):518-524. doi:10.1038/nbt.3423
- Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39(11):1348-1365. doi:10.1038/s41587-021-01108-x


 Sample Submission Guidelines
Sample Submission Guidelines
