Comparative Analysis of DAP-seq and ChIP-seq: Technical Principles, Data Quality, and Applications
In biological research, transcriptional regulatory mapping technologies are crucial for elucidating the regulatory mechanisms of gene expression. With the development of high-throughput sequencing technology, DAP-seq (DNA affinity purification sequencing) and ChIP-seq (chromatin immunoprecipitation sequencing), as two mainstream transcriptional regulatory mapping technologies, have received widespread attention. In this article, DAP-seq and ChIP-seq are compared and analyzed in terms of technical principles, experimental steps, data quality, scope of application, and cost-effectiveness, to provide researchers with a comprehensive understanding and selection guide.
Comparison of Technical Principles and Experimental Procedures
DAP-seq and ChIP-seq differ in their technical principles, with DAP-seq relying on fusion proteins for DNA affinity purification and ChIP-seq on chromatin immunoprecipitation, leading to distinct experimental steps where DAP-seq offers higher automation and reproducibility after fusion protein construction.
Principle of DAP-seq
DAP-seq is a methodology that harnesses the power of DNA affinity purification, in conjunction with high-throughput sequencing technology, to pinpoint the direct binding sites of transcription factors to DNA. The cornerstone of DAP-seq lies in the construction of a fusion protein that encapsulates the DNA-binding domains of the targeted transcription factors. This fusion protein binds efficiently and specifically to the DNA-binding sites. Subsequently, the bound DNA fragments are isolated via an affinity purification step and subjected to rigorous high-throughput sequencing. This comprehensive process allows for the global identification of DNA binding sites for transcription factors, furnishing invaluable data for dissecting transcriptional regulatory networks.
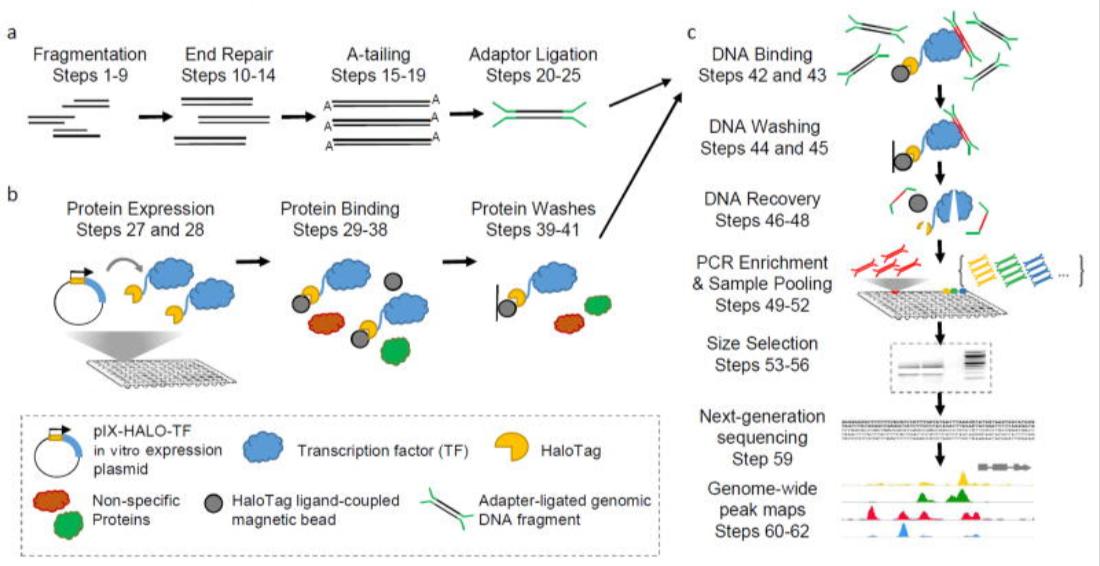 DAP-seq protocol overview (Bartlett et al., 2020)
DAP-seq protocol overview (Bartlett et al., 2020)
Conversely, ChIP-seq employs the technique of chromatin immunoprecipitation to capture DNA fragments that are in intimate contact with specific proteins, such as transcription factors and histone-modifying enzymes. This method utilizes specific antibodies to immunoprecipitate with the target proteins within the cellular milieu, thereby isolating the DNA fragments that are tethered to these proteins. These isolated DNA fragments are then sequenced to unveil the precise binding sites of the protein to the DNA. ChIP-seq is extensively utilized to probe into the protein-DNA interactions that underpin the regulation of gene expression, encompassing transcription factors and histone modifications.
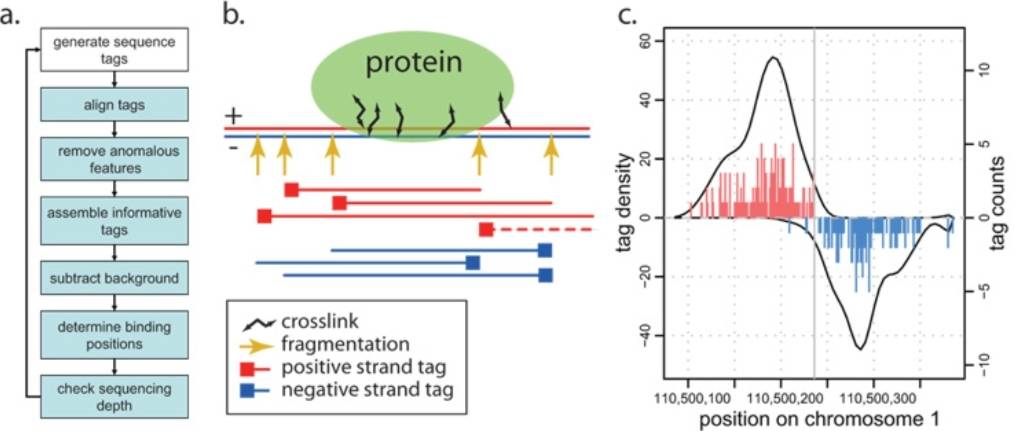 A schematic illustration of ChIP-seq measurements (Kharchenko et al., 2020)
A schematic illustration of ChIP-seq measurements (Kharchenko et al., 2020)
Comparison of experimental steps between DAP-seq and ChIP-seq
There are some differences between DAP-seq and ChIP-seq in terms of experimental steps: DAP-seq requires the construction of fusion proteins by genetic engineering, which increases the complexity of the experiments and the requirements for experimental conditions. However, once the fusion protein has been successfully constructed, the subsequent affinity purification steps are relatively simple and efficient. In contrast, the experimental steps of ChIP-seq include cell fixation, chromatin fragmentation, immunoprecipitation, and DNA extraction, which are not only cumbersome but also susceptible to the influence of cell type, antibody specificity, and other factors. Therefore, in terms of complexity and operability of experimental steps, DAP-seq has higher automation and reproducibility after successful fusion protein construction.
Services you may interested in
Learn More
Data Quality and Resolution Comparison
DAP-seq and ChIP-seq have distinct advantages and limitations in data quality and resolution, with DAP-seq offering higher resolution and efficiency in capturing transcription factor binding sites, as demonstrated in a soybean study that revealed a comprehensive regulatory network.
Data quality
DAP-seq and ChIP-seq have their advantages and disadvantages in terms of data quality. DAP-seq technology can efficiently capture the binding sites of transcription factors by directly exploiting the binding properties of fusion proteins to DNA, thus reducing the interference of background noise. However, data reproducibility can be a challenge as the construction and expression of fusion proteins can affect the natural activity and DNA binding properties of transcription factors. In contrast, ChIP-seq technology relies on the specific binding of antibodies to proteins, which can more realistically reflect the actual state of transcription factors in the cell. However, the specificity and affinity of the antibody directly affect the quality of the data, and differences between different antibodies can lead to less reproducible experimental results.
Resolution comparison
In terms of resolution, the difference between DAP-seq and ChIP-seq is mainly in the accuracy of identification of transcription factor binding sites; DAP-seq technology tends to have higher resolution due to its ability to capture transcription factor binding sites globally and therefore pinpoint specific nucleotide locations. ChIP-seq technology, on the other hand, is limited by the specificity of the antibody and the degree of chromatin fragmentation, and its resolution is relatively low, usually only able to localize larger regions of DNA. However, with continued advances in sequencing technology and optimization of data analysis methods, the resolution of ChIP-seq is gradually improving.
Case study
In a study to explore the transcriptional regulatory network of soybeans in-depth, Jiao et al. systematically analyzed the binding sites of 230 potentially important transcription factors in the soybean genome using DNA affinity purification sequencing (DAP-seq). After rigorous quality control, high-quality data were retained for 148 transcription factors, demonstrating their tight association with soybean growth and development, biotic and abiotic stress responses, and nutrient use. By integrating genome-wide binding maps of these transcription factors with multiple genomics data, the researchers constructed a comprehensive soybean transcription factor-target gene regulatory network that not only covers 2.44 million regulatory relationships between 3188 transcription factors and 51,665 target genes but also provides important clues for revealing candidate transcription factors within genetic loci that are closely associated with multiple agronomic traits. In particular, the researchers successfully predicted important transcription factors that regulate key agronomic traits, such as seed coat color and seed oil content, using the high resolution and reliability of DAP-seq technology. In this study, the DAP-seq dataset demonstrated a high degree of reproducibility, further validating its data quality. In addition, this study is unique in its innovative use of DAP-seq technology to identify transcription factor binding sites in the soybean genome with high precision, providing valuable resources for soybean genetic improvement and molecular breeding. These findings not only deepen our understanding of the transcriptional regulatory network in soybeans but also provide new perspectives and strategies for future crop breeding practices.
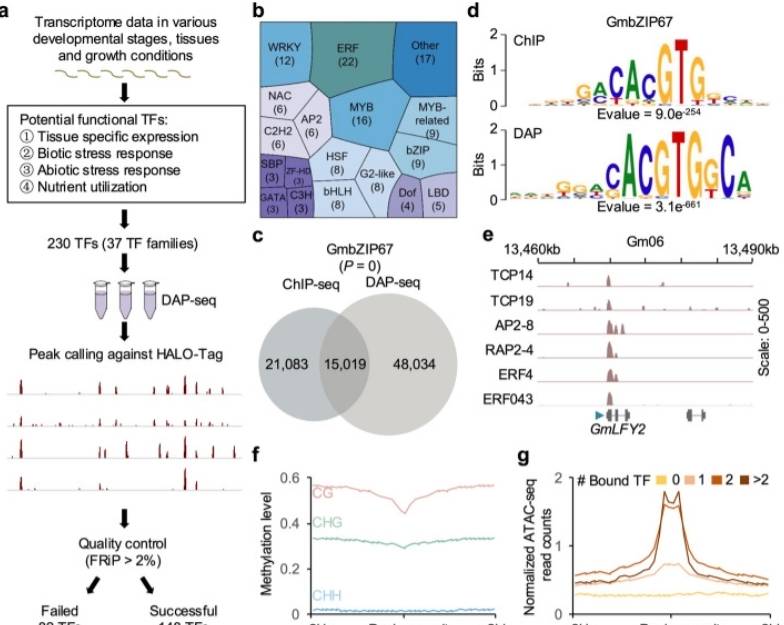 Global identification of TFBSs by DAP-seq (Jiao et al., 2020)
Global identification of TFBSs by DAP-seq (Jiao et al., 2020)
Application Scope and Cost-benefit Analysis
DAP-seq and ChIP-seq have distinct application scopes, with DAP-seq excelling in transcription factor research and regulatory network analysis, while ChIP-seq is better for studying specific protein-DNA interactions; cost-effectiveness and data interpretation also differ, guiding their use in various research scenarios.
Scope of application
In terms of application, DAP-seq and ChIP-seq each have their unique advantages, and DAP-seq is widely used in transcription factor function research and transcriptional regulatory network analysis because of its ability to efficiently and globally identify transcription factor binding sites. In addition, DAP-seq can be used to screen potential transcription factor target genes and provide targets for drug discovery. ChIP-seq, on the other hand, is better suited for studying the interactions between specific proteins and DNA, such as histone modifications, interactions between transcription factors and other regulatory proteins, etc. ChIP-seq can also be used to identify structural changes in chromatin, such as chromatin loops and chromatin domains.
Cost-effectiveness
There are also some differences between DAP-seq and ChIP-seq in terms of cost-effectiveness. The cost of DAP-seq technology is mainly focused on the construction and expression of the fusion proteins, which typically requires a high level of experimental skill and cost investment. However, once the fusion protein has been successfully constructed, the subsequent affinity purification steps and high-throughput sequencing costs are relatively low. In contrast, the costs of ChIP-seq technology are concentrated in the purchase of antibodies, cell culture, chromatin fragmentation, and sequencing. Although the cost of antibodies can be reduced by optimizing experimental conditions and using low-cost alternatives, the overall cost of ChIP-seq technology remains high. In terms of data interpretation, both DAP-seq and ChIP-seq require specialized bioinformatics knowledge and skills, but DAP-seq technology is likely to be simpler and more intuitive in terms of data interpretation due to higher data quality and lower background noise.
Applicable scenarios
Based on the above analysis, we can suggest the application of DAP-seq and ChIP-seq in different research scenarios. For studies that require global identification of transcription factor binding sites and analysis of transcriptional regulatory networks, DAP-seq may be more appropriate. For research that requires the study of specific protein-DNA interactions and changes in chromatin structure, ChIP-seq technology is more appropriate. In addition, researchers need to make comprehensive considerations based on experimental conditions, cost budget, and data interpretation capabilities to choose the most appropriate transcriptional regulatory mapping technology.
Technology Challenges and Future Trends
DAP-seq and ChIP-seq face technical challenges in protein fusion expression and antibody specificity, but future trends indicate improved resolution, cost reduction, and integration with other histological data for comprehensive gene expression analysis.
Technical challenges
DAP-seq and ChIP-seq technologies still face some challenges in practical applications. For DAP-seq, the construction and expression of fusion proteins are key issues. How to ensure that the fusion proteins are stably expressed in cells and retain their natural activity and DNA-binding properties is a focus of current research. In addition, DAP-seq technology requires further optimization of experimental conditions and data analysis methods to improve data quality and resolution. For ChIP-seq technology, antibody specificity and affinity are key factors affecting data quality. How to screen and optimize antibodies to reduce background noise and false positives is another focus of current research. In addition, ChIP-seq technology needs to overcome issues such as cell type restriction and incomplete chromatin fragmentation to improve its application range and resolution.
Future trends
Looking at the future direction of transcriptional regulatory mapping technology, we can foresee the following trends: first, improving resolution. As sequencing technology continues to advance and data analysis methods are optimized, the resolution of DAP-seq and ChIP-seq technologies will continue to improve, enabling more precise identification of transcription factors and DNA binding sites. Second, cost reduction. By optimizing experimental conditions, using low-cost alternatives, and improving the efficiency of data interpretation, the cost of DAP-seq and ChIP-seq technologies will be further reduced, making them more popular and easier to use. Third, the combination of other histological data. The future transcriptional mapping technology will pay more attention to the integration and analysis of other histological data (e.g. genome, transcriptome, proteome, etc.) to comprehensively analyze the regulatory mechanism of gene expression.
Selection and Application of DAP-seq/ChIP-seq
DAP-seq and ChIP-seq have unique advantages and researchers should choose the most suitable technology based on research goals, conditions, and other factors to elucidate gene expression regulatory mechanisms and advance biological and medical research.
For a more detailed comparison of DAP-seq and ChIP-seq, the following table summarizes their key characteristics and differences:
| DAP-seq | ChIP-seq | |
|---|---|---|
| Technical Principle | Uses fusion proteins for DNA purification | Captures DNA-protein complexes via immunoprecipitation |
| Experimental Steps | Fusion protein construction, purification, sequencing | Cell fixation, chromatin fragmentation, immunoprecipitation, sequencing |
| Complexity | Complex in protein fusion construction, simple purification | Cumbersome, influenced by cell type and antibody |
| Automation & Reproducibility | High automation, good reproducibility after fusion construction | Lower reproducibility due to multiple factors |
| Data Quality | Efficient capture of TFBS, low background noise | Reflects in-cell TF state, influenced by antibody quality |
| Resolution | Higher, pinpoints TFBS to specific nucleotides | Lower, localizes larger DNA regions |
| Applications | TF function research, regulatory network analysis | Protein-DNA interactions, histone modifications |
| Cost-effectiveness | High initial fusion protein costs, low subsequent costs | High antibody and cell culture costs |
| Data Interpretation | Relatively simple due to high data quality | Requires specialized bioinformatics skills |
| Challenges | Fusion protein stability, data analysis optimization | Antibody specificity, chromatin fragmentation |
Conclusion
In summary, DAP-seq and ChIP-seq, as two mainstream technologies for mapping transcriptional regulation, have their advantages and disadvantages in terms of technical principles, experimental steps, data quality, scope of application, cost-effectiveness, etc. DAP-seq technology, with its high efficiency and global characteristics, has unique advantages in identifying transcription factor binding sites and resolving transcriptional regulatory networks, etc., while ChIP-seq technology, with its high specificity and wide applicability, is widely used in studying specific protein-DNA interactions and chromatin structure changes. Researchers should select the most appropriate transcriptional regulation mapping technology according to the purpose and conditions of the study, taking into account various factors. In the future, with the continuous progress of technology and the expansion of application, DAP-seq and ChIP-seq technologies will play an even more important role in elucidating the regulatory mechanism of gene expression and promoting biological research and medical development.
Recommendations
Researchers should fully consider factors such as research purpose, experimental conditions, cost budget, and data interpretation capabilities when choosing between DAP-seq and ChIP-seq technologies. For studies requiring the global identification of transcription factor binding sites, DAP-seq may be more appropriate, whereas, for studies requiring the interaction of specific proteins with DNA, ChIP-seq may be more appropriate. In addition, researchers may also consider combining other histological data for comprehensive analysis to more fully elucidate the regulatory mechanism of gene expression. Through the rational selection and application of transcriptional mapping technologies, researchers will be able to better understand the complexity and diversity of gene expression regulation and make greater contributions to biological research and medical development.
References:
- Bartlett A, O'Malley RC., et al. "Mapping genome-wide transcription-factor binding sites using DAP-seq". Nat Protoc. 2017 Aug;12(8):1659-1672.https://doi.org/10.1038/nprot.2017.055
- Kharchenko PV, Tolstorukov MY., et al. "Design and analysis of ChIP-seq experiments for DNA-binding proteins." Nat Biotechnol. 2008 Dec;26(12):1351-9. https://doi.org/10.1038/nbt.1508
- Jiao W, Wang M., et al. "Transcriptional regulatory network reveals key transcription factors for regulating agronomic traits in soybean." Genome Biol. 2024 Dec 18;25(1):313. https://doi.org/10.1186/s13059-024-03454-w


 Sample Submission Guidelines
Sample Submission Guidelines
