Expanding Horizons: Diverse Applications of DAP-Seq
Gene regulation represents the cornerstone of biological complexity, governing the spatiotemporal precision of cellular differentiation, developmental patterning, and environmental adaptation. Understanding how transcription factors (TFs) bind to DNA and regulate target genes is key to unravelling the complexity of gene networks. In this context, DNA affinity purification sequencing (DAP-Seq), with its high-throughput and antibody-independent features, has become a revolutionary tool for genome-wide resolution of protein-DNA interactions. DAP-Seq accurately identifies transcription factor binding sites (TFBSs) through in vitro expression of TFs and binding to genomic DNA libraries in combination with high-throughput sequencing technology. DAP-Seq offers unprecedented opportunities for genomic studies in model and non-model species.
DAP-Seq Decoding Plant-Environment Adaptation
Plants sense and respond to environmental changes through molecular mechanisms, and transcription factors are the central regulators of this process. DAP-Seq is particularly advantageous for studies of plant-environment interactions, especially in non-model species.
A study of nitrogen use efficiency (NUE) in rice: scientists used DAP-Seq technology to discover that the transcription factor OsGATA8 balances nitrogen uptake and tiller formation by repressing the expression of the ammonium transporter protein gene OsAMT3.2 and the tiller repressor OsTCP19, thereby optimising crop yield.
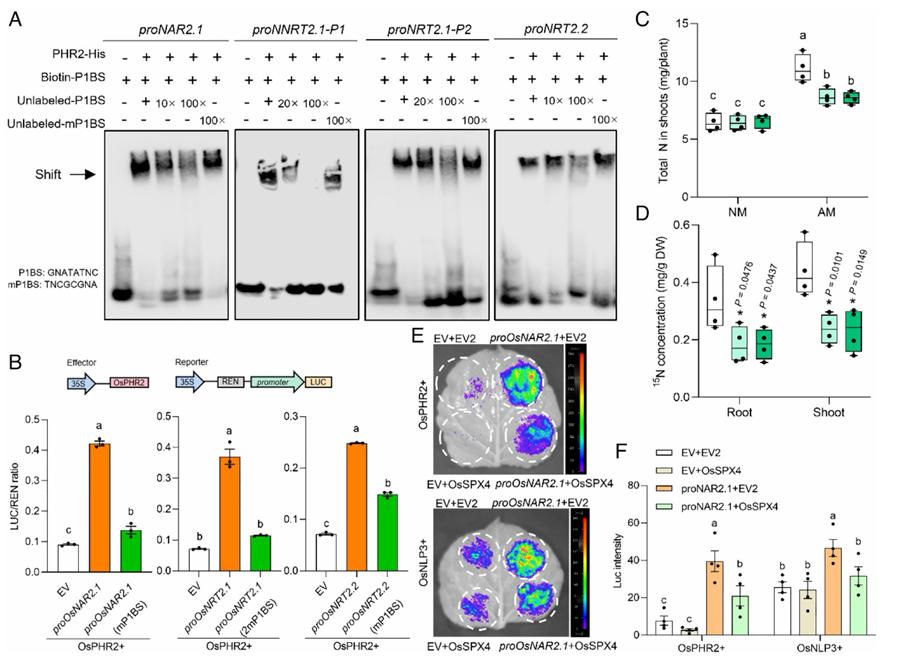 OsPHR2 regulates mycorrhizal NO3− uptake via directly modulating OsNAR2.1 and OsRT2.1/2.2 (Wang et al., 2025)
OsPHR2 regulates mycorrhizal NO3− uptake via directly modulating OsNAR2.1 and OsRT2.1/2.2 (Wang et al., 2025)
A study of disease resistance mechanisms in citrus: DAP-Seq combined with transcriptome analysis revealed that CsWRKY33 enhances fruit disease resistance by regulating oxidative stress genes and pathways. Such studies not only deepen the understanding of plant adaptation to adversity, but also provide targets for molecular design breeding. Innovative experimental designs, such as cross-species comparisons (e.g. wheat vs. Arabidopsis thaliana) or dynamic analyses at multiple time points, further expand the dimensions of the application of DAP-Seq in ecological adaptation studies.
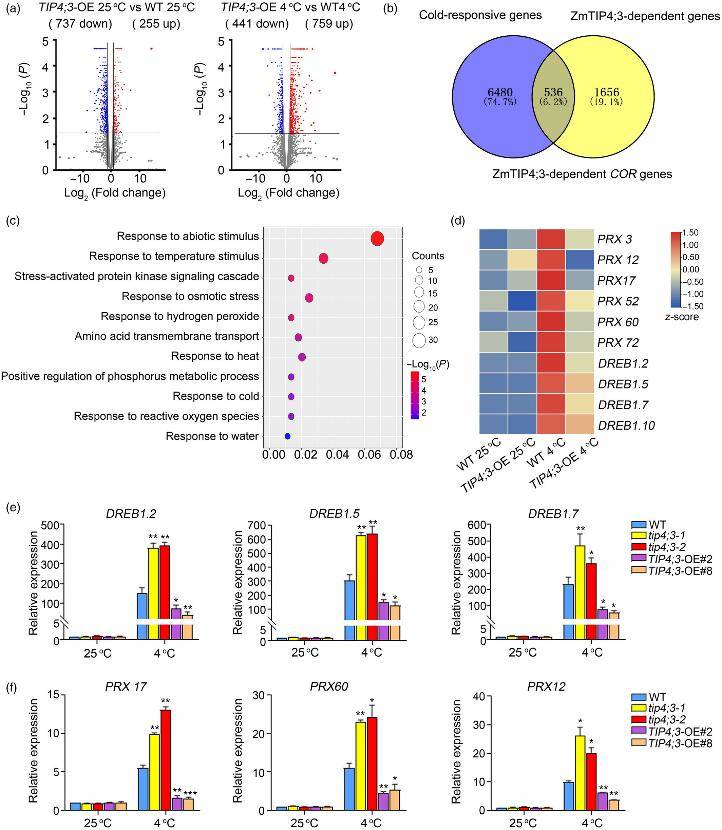 Transcriptome analysis of TIP4;3-dependent genes (Zeng et al., 2024)
Transcriptome analysis of TIP4;3-dependent genes (Zeng et al., 2024)
Services you may interested in
Learn More
Applications of DAP-Seq in Epigenetic Research
Epigenetic studies focus on the dynamic regulatory mechanisms of chromatin state and gene expression under conditions of unaltered DNA sequence. By designing in vitro experiments, DAP-Seq is able to directly capture information on the interactions between transcription factors and DNA, and thus indirectly reflect the effects of epigenetic modifications on gene activity. In plants, for example, chromatin accessibility is usually closely linked to the binding of specific transcription factors. DAP-Seq not only identifies open chromatin regions, but also reveals the regulatory effects of epigenetic marks (e.g. histone modifications) on transcription factor recruitment by analysing the distribution of binding sites. Compared to traditional ChIP-Seq, DAP-Seq does not require antibodies, thus avoiding the problem of false negatives due to insufficient antibody specificity, while achieving single base resolution.
A study of heat tolerance in apple: DAP-Seq successfully identified the promoter region of the heat shock factor gene bound by the MdWRKY75 transcription factor under heat stress, revealing the molecular link between epigenetic regulation and basal heat tolerance. In addition, DAP-Seq data can be complemented by ATAC-Seq (chromatin openness analysis) to construct a complete epigenetic regulatory network from chromatin structure to transcription factor binding.
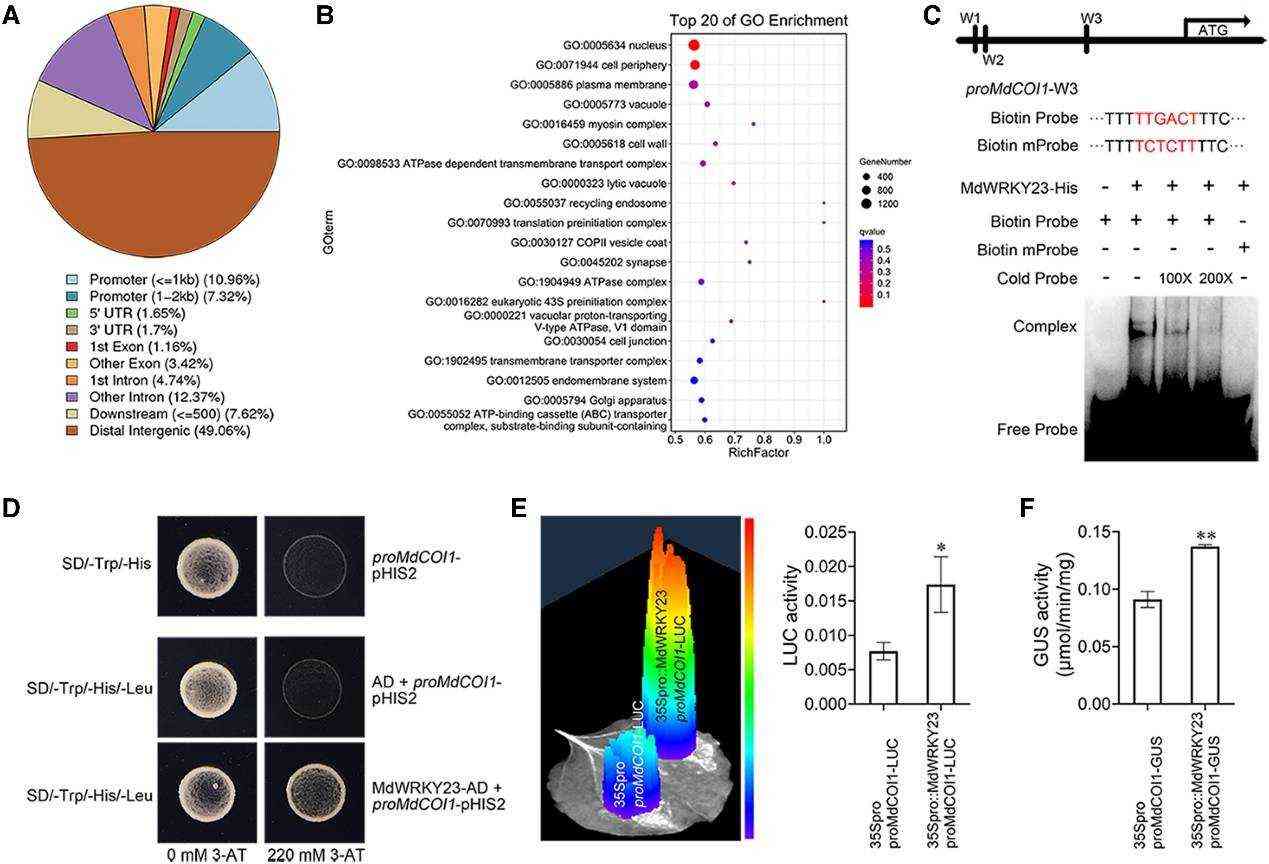 MdWRKY23 transcriptionally activates MdCOI1 expression to regulate the JA signaling pathway (Han et al., 2024)
MdWRKY23 transcriptionally activates MdCOI1 expression to regulate the JA signaling pathway (Han et al., 2024)
A study of triterpenoid regulation in Ganoderma lingzhi: DAP-Seq identified the SREBP binding motif (5'-GRVGRVGRVGR-3') and 1,532 target genes enriched in mevalonate (MVA) and ergosterol pathways. Multi-omics analysis revealed SREBP overexpression upregulates key enzymes (e.g., HMGS, MVD) and enhances ganoderic acid production by 36.15%, while pharmacological inhibition with fatostatin reversed these effects, confirming SREBP's negative feedback regulation. Notably, SREBP also activated glycerophospholipid metabolism genes (g1208, g4309), linking lipid homeostasis to secondary metabolite synthesis. This study provides the first evidence of SREBP's non-animal regulatory role and proposes transcriptional engineering strategies to optimize fungal medicinal compound yields.
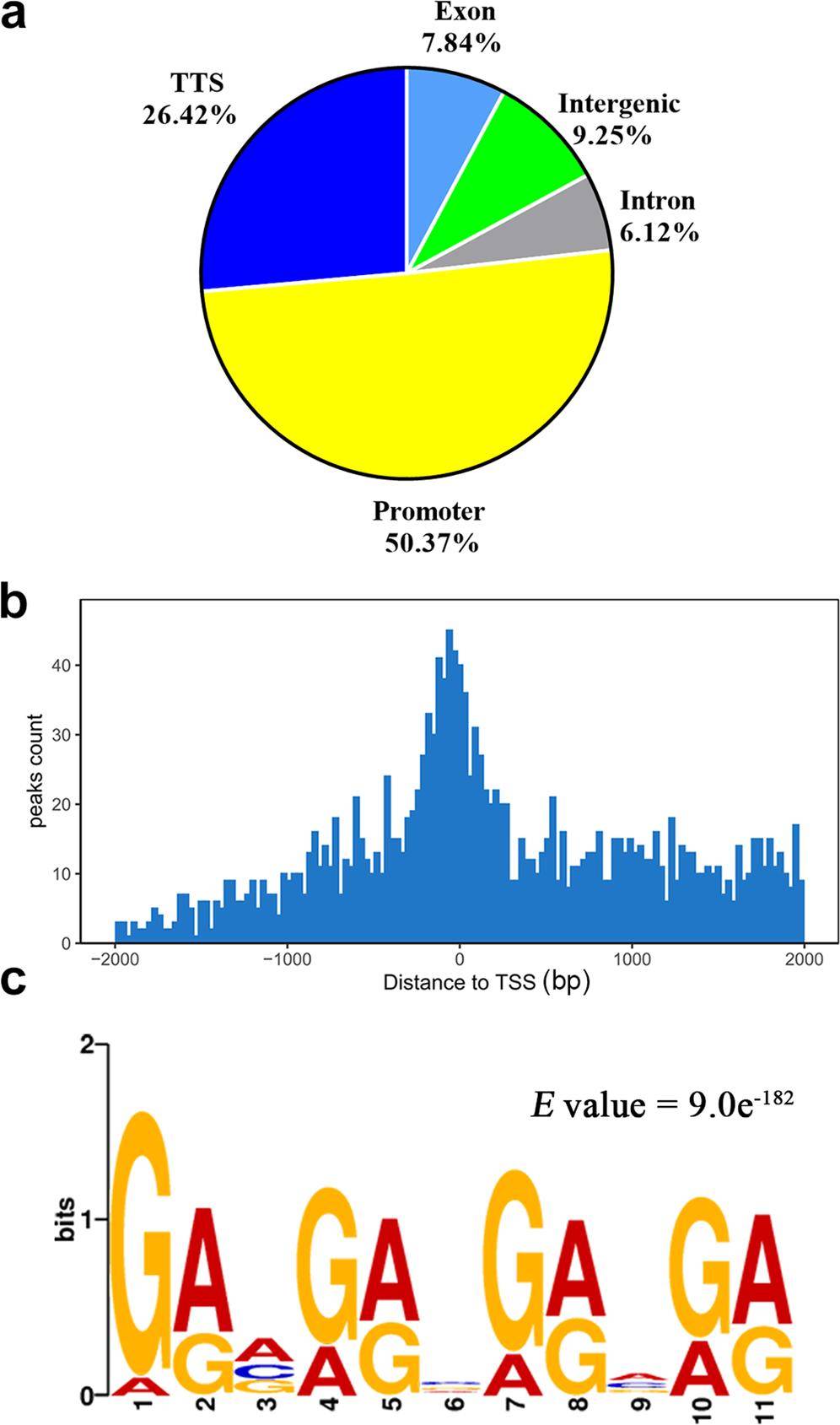 The bHLH-zip transcription factor SREBP regulates triterpenoid and lipid metabolisms in the medicinal fungus Ganoderma lingzhi (Liu et al., 2023)
The bHLH-zip transcription factor SREBP regulates triterpenoid and lipid metabolisms in the medicinal fungus Ganoderma lingzhi (Liu et al., 2023)
A study of brassinosteroid (BR) signaling in cotton fiber: identified GhBES1.4 as a master regulator through DAP-Seq profiling of 1,531 direct targets, including BR biosynthesis genes (GhCPD, GhCYP90D1) and cell elongation drivers (GhBRL). Functional validation showed GhBES1.4 overexpression increased fiber length by activating fatty acid metabolism and ethylene signaling, while silencing impaired elongation. Integration with RNA-seq and GWAS data prioritized seven key genes (e.g., GhCYP84A1) that enhance fiber cell expansion through E-box motif binding. BR biosynthesis inhibitor (BRZ) treatments further confirmed the BR-GhBES1.4 regulatory dependency. This work establishes a hierarchical transcriptional network and highlights GhBES1.4 as a prime target for molecular breeding to improve cotton fiber quality and agricultural sustainability.
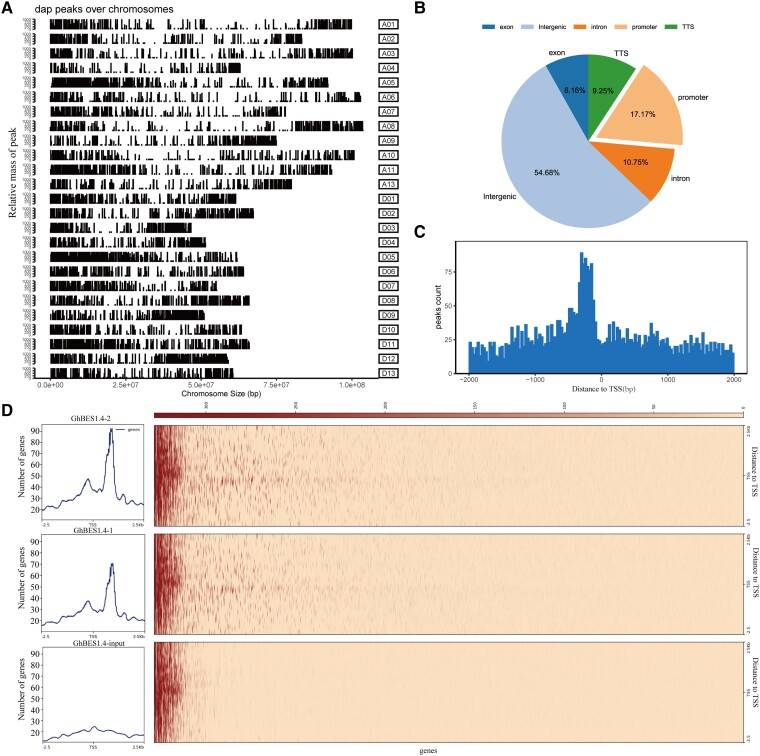 Identification of direct target genes of GhBES1.4 using DAP-seq (Liu et al., 2023)
Identification of direct target genes of GhBES1.4 using DAP-seq (Liu et al., 2023)
DAP-Seq-Driven Innovations in Synthetic Biology
Synthetic biology aims to modify biological systems through engineering design, and precise gene regulatory elements are at the core of the construction of artificial pathways or biosensors. DAP-Seq provides data support for the rational design of synthetic biology components by providing genome-wide binding maps of natural transcription factors. For example, in the construction of microbial cell factories, researchers have used DAP-Seq data to optimise promoter sequences and increase the expression efficiency of exogenous genes. In plant synthetic biology, DAP-Seq has been used to design artificial transcription factors for targeted regulation of the biosynthesis of secondary metabolites such as medicinal alkaloids. In a breakthrough case, a DAP-Seq-guided CRISPR-activated system successfully increased the yield of tanshinones in Salvia divinorum. In addition, DAP-Seq helped assess the stability of synthetic gene circuits and reduced the metabolic burden of engineered strains by predicting off-target binding of transcription factors to non-target DNA.
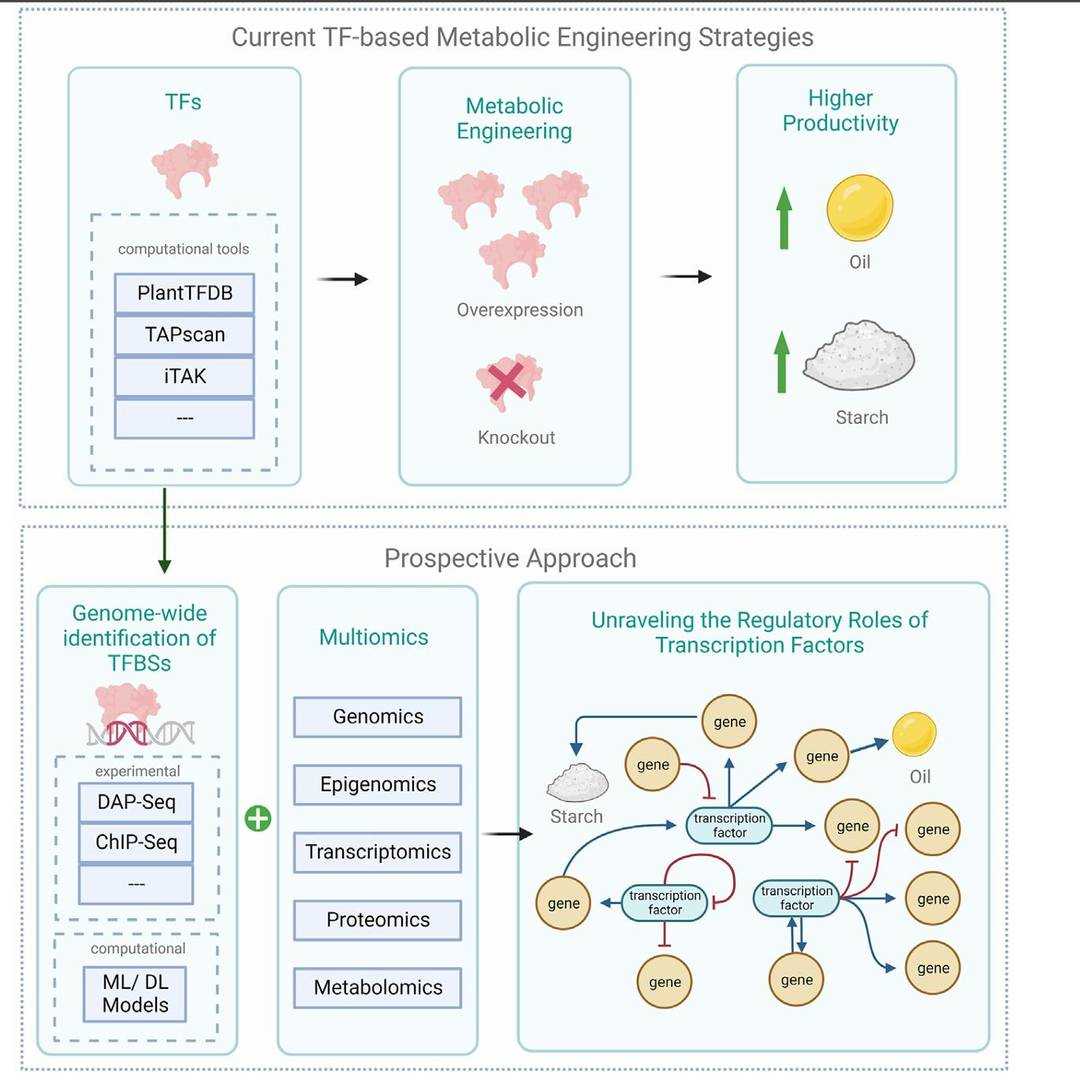 Schematic of current transcription factor-based engineering strategies in microalgae alongside prospective approaches (Gupta et al., 2024)
Schematic of current transcription factor-based engineering strategies in microalgae alongside prospective approaches (Gupta et al., 2024)
DAP's dual function as both a precursor for storage molecules and a regulator of redox balance, which is essential for maintaining cellular homeostasis under stress conditions. Through targeted overexpression of DAP-metabolizing enzymes, such as glycerol-3-phosphate dehydrogenase, the engineered algal strains exhibited improved tolerance to environmental stressors like high salinity and nutrient limitation. This resilience directly translates to scalable cultivation systems, reducing reliance on freshwater resources and costly growth media. Furthermore, the study underscores DAP's role in synchronizing carbon fixation with downstream biosynthesis, enabling dynamic control over metabolic pathways via light-responsive genetic circuits.
Challenges and Future of DAP-Seq
While DAP-Seq offers high-throughput identification of transcription factor binding sites (TFBSs), its in vitro framework faces critical gaps: (1) inability to replicate chromatin dynamics (e.g., DNA methylation/histone modification-dependent TF binding), (2) blindness to heterodimer interactions and (3) noise from low-abundance TFs or repetitive sequences. To overcome these, next-gen adaptations like dual-tagged DAP-Seq (dDAP-Seq) now decode genome-wide heterodimer co-binding, while single-cell spatial transcriptomics maps spatiotemporal TF activity in contexts like hepatocellular carcinoma (e.g., MYC oncogene dynamics). AI-driven motif prediction tools further refine experimental design, boosting efficiency in applications such as tanshinone biosynthesis pathway engineering.
Future advancements prioritize epigenetically preserved DNA libraries (e.g., dissecting OsNLP3-mediated nitrogen uptake under stress in rice), cross-omics data fusion (e.g., HNF4α's dual metabolic control in liver cancer via glycolysis/OXPHOS crosstalk), and 3D genome integration for dynamic processes like organogenesis. By merging physiological relevance with multi-scale resolution, DAP-Seq is evolving into a unique, cross-disciplinary toolkit, addressing both plant stress adaptation and cancer mechanisms while bypassing traditional ChIP-Seq limitations. This innovation-driven reinvention positions DAP-Seq as a Google-search standout for precision transcriptional regulation studies.
Conclusion
Since its launch in 2016, DAP-Seq has become a key tool for genomics research. In plants, it has revealed the molecular mechanisms by which citrus CsWRKY33 enhances disease resistance through the oxidative stress pathway and rice OsGATA8 coordinates nitrogen use and tiller formation. In epigenetics, the technology combined with ATAC-Seq to construct an association model of chromatin openness and transcription factor binding.
The continued innovation of DAP-Seq technology will bring genomics into the era of multi-dimensional resolution. The combination of single-cell spatio-temporal genomics and artificial intelligence-driven predictive models is expected to provide mechanistic breakthroughs in complex biological problems such as stem cell differentiation or ecological adaptive evolution. Although the physiological relevance of in vitro experiments still needs to be compensated by organoid models or in situ validation, the integration of interdisciplinary approaches (e.g. synthetic biology and systems biology) will make them more applicable. As shown by the breakthrough of dDAP-seq in dimer research, the deep coupling of technological innovation and biological problems will surely lead to more groundbreaking discoveries in the life sciences.
References:
- Wang S, Ye H, Yang C, Zhang Y, Pu J, Ren Y, Xie K, Wang L, Zeng D, He H, Ji H, Herrera-Estrella LR, Xu G, Chen A. "OsNLP3 and OsPHR2 orchestrate direct and mycorrhizal pathways for nitrate uptake by regulating NAR2.1-NRT2s complexes in rice." Proc Natl Acad Sci U S A. 2025 122(8):e2416345122. https://doi.org/10.1073/pnas.2416345122
- Zeng R, Zhang X, Song G, Lv Q, Li M, Fu D, Zhang Z, Gao L, Zhang S, Yang X, Tian F, Yang S, Shi Y. "Genetic variation in the aquaporin TONOPLAST INTRINSIC PROTEIN 4;3 modulates maize cold tolerance." Plant Biotechnol J. 2024 22(11):3037-3050. https://doi.org/10.1111/pbi.14426
- Han P, Wang C, Li F, Li M, Nie J, Xu M, Feng H, Xu L, Jiang C, Guan Q, Huang L. "Valsa mali PR1-like protein modulates an apple valine-glutamine protein to suppress JA signaling-mediated immunity." Plant Physiol. 2024 194(4):2755-2770 https://doi.org/10.1093/plphys/kiae020
- Liu L, Chen G, Li S, Gu Y, Lu L, Qanmber G, Mendu V, Liu Z, Li F, Yang Z. "A brassinosteroid transcriptional regulatory network participates in regulating fiber elongation in cotton." Plant Physiol. 2023 191(3):1985-2000. https://doi.org/10.1093/plphys/kiac590
- Liu YN, Wu FY, Tian RY, Shi YX, Xu ZQ, Liu JY, Huang J, Xue FF, Liu BY, Liu GQ. "The bHLH-zip transcription factor SREBP regulates triterpenoid and lipid metabolisms in the medicinal fungus Ganoderma lingzhi." Commun Biol. 2023 6(1):1. https://doi.org/10.1038/s42003-022-04154-6
- Gupta A, Kang K, Pathania R, Saxton L, Saucedo B, Malik A, Torres-Tiji Y, Diaz CJ, Dutra Molino JV, Mayfield SP. "Harnessing genetic engineering to drive economic bioproduct production in algae." Front Bioeng Biotechnol. 2024 12:1350722. https://doi.org/10.3389/fbioe.2024.1350722


 Sample Submission Guidelines
Sample Submission Guidelines
