As a key phenomenon in the field of molecular biology, reverse transcription subverts the traditional "central rule" cognition and greatly expands people’s understanding of the law of genetic information transmission. This process refers to the process of synthesizing DNA with RNA as a template under the catalysis of reverse transcriptase, which breaks the one-way flow mode that genetic information can only flow from DNA to RNA.
Nowadays, reverse transcription plays a crucial role not only in biological processes such as virus replication, cell differentiation, and development, but also in biotechnology applications including genetic engineering, disease diagnosis, and therapy. It forms the foundation of transcriptome sequencing (RNA-Seq), enabling comprehensive analysis of gene expression in various physiological and pathological states.
This paper discusses the process of reverse transcription, including its definition, structure and function of reverse transcriptase, expounds its application, analyzes the challenges and prospects.
What is Reverse Transcription
In the traditional central law of biology, the flow direction of genetic information is from DNA to RNA and then to protein. However, the discovery of Reverse Transcription breaks this inherent cognition, which reveals that genetic information can be transmitted from RNA to DNA in reverse, bringing a new perspective and field to the research of life science.
Reverse transcription refers to the process of synthesizing DNA with RNA as a template, which is opposite to transcription (synthesizing RNA with DNA as a template), so it is called reverse transcription. Reverse transcription was first discovered independently by American scientists Howard Temin and David Baltimore in 1970. When studying RNA tumor viruses, they found that these viruses can reverse transcribe their RNA genomes into DNA and integrate them into the genome of host cells, thus causing cancer of cells. This discovery not only subverted the traditional concept of genetic information flow at that time, but also laid an important foundation for later genetic engineering and molecular biology research.
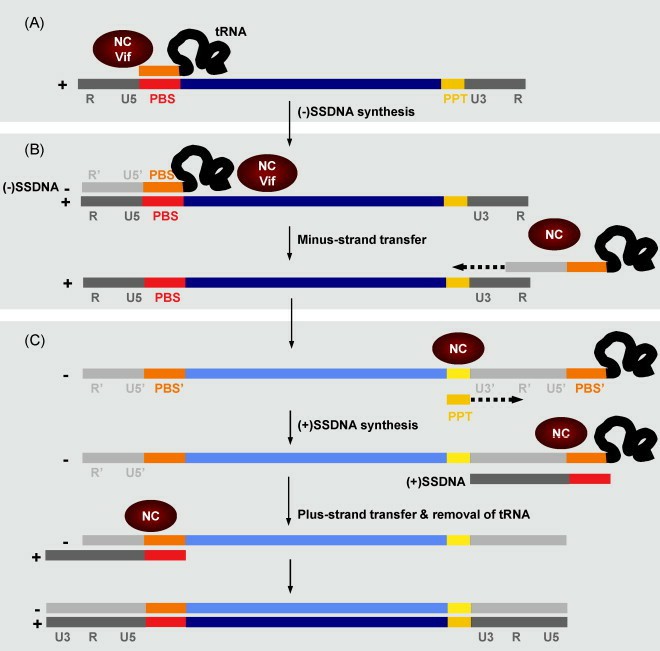
Representation of the key steps in retrovirus replication (Zúñiga et al., 2008)
The Structure of Reverse Transcriptase
As a unique enzyme, reverse transcriptase can catalyze the process of synthesizing DNA with RNA as a template, and realize the reverse flow of genetic information from RNA to DNA. Exploring the structure and function of reverse transcriptase will not only help us reveal the mystery of life inheritance and evolution from the molecular level, but also provide a solid theoretical basis for developing therapeutic strategies for reverse transcription-related diseases and promoting the innovative development of biotechnology. The basic structural composition, the functions of each domain and the structural characteristics of reverse transcriptase from different sources will be introduced in detail below.
Structure of Reverse Transcriptase
Reverse transcriptase usually has a complex three-dimensional structure, and its overall shape can be vividly compared to a "right hand", which is a common structural feature of many members of DNA polymerase family. This "right-handed" structure contains three main domains, corresponding to the palm, finger and thumb regions respectively, and these domains work together to complete various functions in the reverse transcription process.
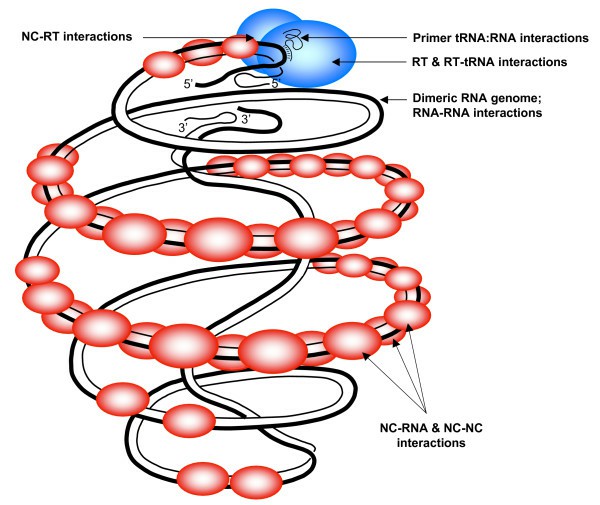
A scheme of the HIV-1 replication complex (Mougel et al., 2009)
Palm Domain
Palm domain is the core region of reverse transcriptase, which contains the active center that catalyzes reverse transcription reaction. The active center is rich in some specific amino acid residues, such as aspartic acid (Asp), which can combine with metal ions (usually magnesium ions) to form the microenvironment required for catalytic activity. In the process of reverse transcription, metal ions can promote the base pairing between substrate and template RNA, and catalyze the formation of phosphodiester bond, thus realizing the extension of DNA chain.
Finger Domain
Finger domain is located at the N-terminal of reverse transcriptase, and its main function is to recognize and bind primer-template complex. This domain has a specific amino acid sequence and spatial conformation, and can form accurate interaction with RNA or DNA templates and primers. During the binding process, the conformation of the finger domain will change, so that the primer-template complex will be fixed near the active center to prepare for the subsequent polymerization reaction.
Thumb Domain
The thumb domain is located opposite to the palm domain, which mainly plays the role of stabilizing the primer-template complex and regulating the polymerization process. In the process of polymerization, thumb domain will interact with DNA or RNA chain, and change its conformation to adapt to nucleic acid chains with different lengths and structures to ensure the smooth progress of polymerization. At the same time, the thumb domain can also cooperate with other domains to regulate the activity and catalytic efficiency of reverse transcriptase.
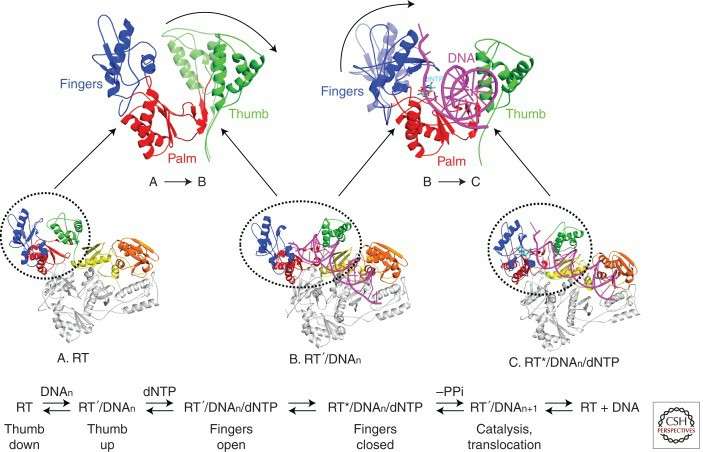
Structural changes in RT that occur during polymerization (Hu et al., 2012)
Different Reverse Transcriptases Comparison
Virus-derived reverse transcriptase: Take the reverse transcriptase of human immunodeficiency virus (HIV) as an example. It is a heterodimer composed of two subunits, p66 and p51. p66 subunit contains complete RNA-dependent DNA polymerase domain, RNase H domain and DNA-dependent DNA polymerase domain. The p51 subunit is the product of removing RNase H domain from p66 subunit by protease cleavage. This heterodimer structure makes HIV reverse transcriptase have unique catalytic activity and regulatory mechanism. The two subunits interact with each other in space to form a specific three-dimensional structure and complete the reverse transcription process together. Among them, the p66 subunit is mainly responsible for catalyzing the reaction, while the p51 subunit may play a role in stabilizing the structure and regulating the activity.
Reverse transcriptase in telomerase: Telomerase is a special reverse transcriptase, and its structure is different from that of retrovirus. Telomerase is composed of protein subunit and RNA subunit, in which protein subunit has reverse transcriptase activity. The RNA subunit of telomerase not only serves as a template to guide the synthesis of telomere DNA, but also interacts with protein subunits to form the active center of telomerase. This unique structure enables telomerase to specifically recognize and prolong telomere sequences at the end of chromosomes, and maintain the stability of chromosomes and the proliferation ability of cells.
How Does Reverse Transcriptase Function
Reverse transcriptase is a key enzyme in the field of molecular biology. Understanding the activity of reverse transcriptase is of great significance to the study of virus pathogenesis, the development of biotechnology and the treatment of related diseases.
RNA-dependent DNA Polymerase Activity
One of the core functions of reverse transcriptase is to use RNA as a template to synthesize a complementary DNA strand, that is, the first strand of cDNA, according to the principle of base complementary pairing. In this process, primers played a key initial role. Common primer types are oligo (dT) primers, random primers and gene-specific primers. When oligo (dT) primer is used, it will combine with the poly-A tail of mRNA to provide the site for reverse transcriptase to start synthesis. Reverse transcriptase starts from the 3′-hydroxyl end of the primer, adds deoxynucleotides one by one, and moves along the 3’→5′ direction of the RNA template to synthesize the first cDNA strand in the 5’→3′ direction.
Rnase H Activity
In the process of reverse transcription, RNA-DNA hybrids will be formed with the synthesis of the first strand of cDNA. The RNase H activity of reverse transcriptase can specifically degrade the RNA chain in this hybrid. It will cut the RNA chain into small fragments, creating conditions for the subsequent synthesis of the second strand of cDNA. The activity of RNase H is very important to maintain the normal reverse transcription process. If the RNA in RNA-DNA hybrid is not degraded in time, it may affect the subsequent reaction steps and lead to the reduction of reverse transcription efficiency. At the same time, it also helps to remove redundant RNA templates and avoid their interference with subsequent experiments or biological processes.
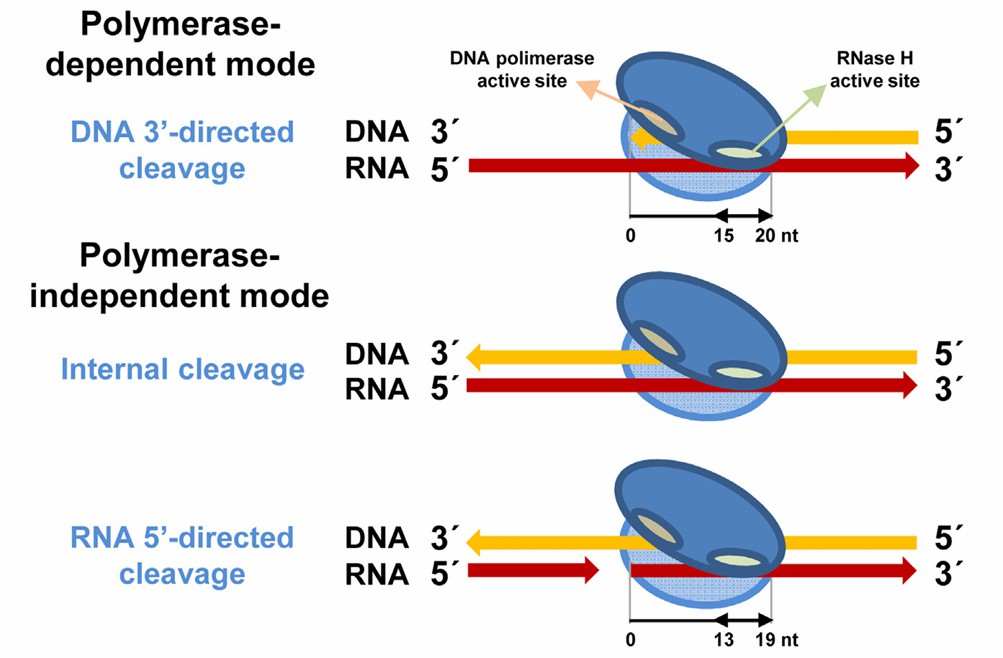
Retroviral RT binding modes to RNA/DNA hybrids and effects on RNase H cleavage (Menéndez-Arias et al., 2017)
DNA-dependent DNA Polymerase Activity
After RNA in RNA-DNA hybrid is degraded, reverse transcriptase can use its DNA-dependent DNA polymerase activity to synthesize a complementary second strand with the first strand of the synthesized cDNA as a template, thus forming a double-stranded cDNA. This process is similar to DNA polymerization in the process of DNA replication. Reverse transcriptase will move along the 3’→5′ direction of the first strand of cDNA to synthesize the second strand in the 5’→3′ direction.
Applications of Reverse Transcription
From helping to accurately diagnose diseases, to opening up a new path for gene therapy, and then to playing a central role in the biopharmaceutical industry, the application scope of reverse transcription continues to expand, and it continues to provide innovative solutions for solving many complex problems, profoundly affecting and promoting the rapid development of life science-related fields.
Gene Engineering
Construction of cDNA library: Construction of cDNA library is one of the important applications of reverse transcription technology in genetic engineering. By extracting mRNA from cells or tissues, using reverse transcription technology to synthesize cDNA, and then cloning the cDNA into a suitable vector and introducing it into host cells, a library containing a large number of cDNA clones is formed. cDNA library can be used for gene cloning, gene expression analysis and protein omics research.
Study on gene expression analysis and regulation: RT-qPCR is a commonly used gene expression analysis technique. It converts mRNA into cDNA by reverse transcription, and then uses PCR technology to amplify and quantify the cDNA, so as to accurately determine the expression level of specific genes in different tissues, different development stages or different treatment conditions. By comparing the differences of gene expression in different samples, we can reveal the regulation mechanism of gene expression and find the key genes related to the occurrence and development of diseases.
Medical Diagnosis
Diagnosis and monitoring of virus infection: Reverse transcription PCR technology has important application in the diagnosis and monitoring of virus infection. Many viruses (such as HIV, hepatitis B virus, hepatitis C virus, etc.) have RNA genomes. The viral RNA is converted into cDNA by reverse transcription, and then the cDNA is amplified and detected by PCR technology, which can quickly and accurately diagnose viral infection. In addition, by monitoring the change of viral load, we can evaluate the effect of antiviral treatment and guide the adjustment of clinical treatment scheme.
Tumor diagnosis and prognosis evaluation: The gene expression pattern of tumor cells is different from that of normal cells. Detecting the expression level of specific mRNA in tumor cells by reverse transcription technology can provide an important basis for tumor diagnosis, typing and prognosis evaluation.
Evolutionary Biology
Analysis of species evolutionary relationship: Reverse transcription technology can be used to analyze the differences of RNA sequences in different species, and then infer the evolutionary relationship between species. By reverse transcription and sequencing the mRNA of specific genes or genomic regions of different species and comparing their cDNA sequences, we can construct the phylogenetic tree of species and reveal the evolutionary process and genetic relationship of species.
Study on molecular evolution mechanism: At the molecular level, gene mutation and recombination during reverse transcription provide important clues for studying molecular evolution mechanism. Through the analysis of reverse transcription products, we can study the origin, evolution and functional evolution of genes, and understand the transmission and variation law of genetic information in the process of evolution.
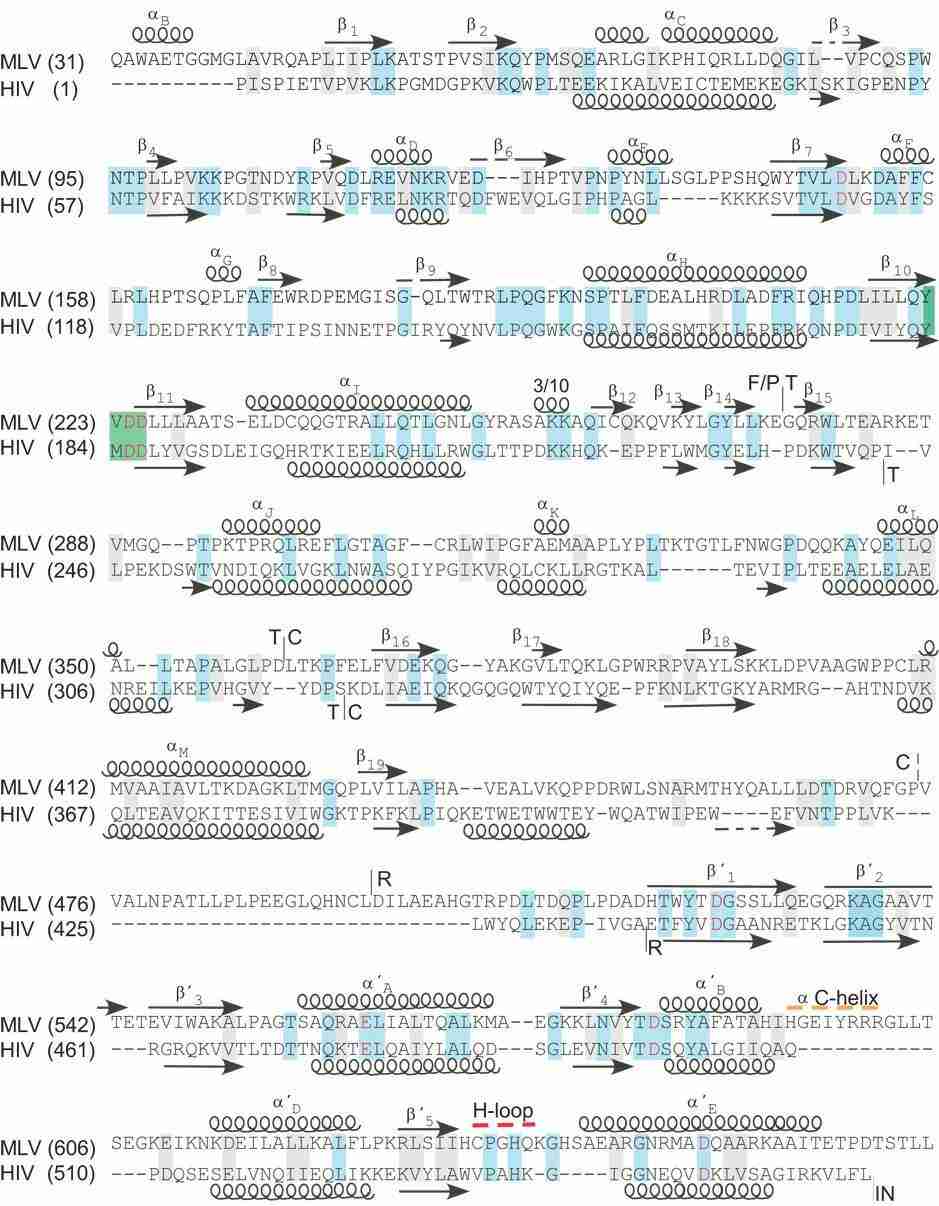
Sequence alignment and structural motifs of the MoMLV and HIV-1 RTs (Coté et al., 2008)
By combining the cDNA generated by reverse transcription with the next generation sequencing technology (NGS), researchers can comprehensively analyze the composition and dynamic changes of transcriptome. For example, RNA sequencing (RNA-seq) technology uses reverse transcription to convert RNA into cDNA, and then obtains the quantitative and qualitative data of the whole transcript through sequencing. This method can not only detect the expression level of genes, but also discover new transcripts, alternative splicing events and functions of non-coding RNA.
Challenges and Prospects in Reverse Transcription
As a unique biological process, reverse transcription plays a key role in virology, genetics and modern biotechnology. It not only breaks the traditional direction of genetic information flow, but also provides new ideas and methods for gene therapy and disease diagnosis. However, with the deepening of research, reverse transcription research is also facing many challenges. At the same time, new technologies and ideas have brought broad prospects for its future development.
Challenges of Reverse Transcription
Performance limitation of reverse transcriptase: During reverse transcription, reverse transcriptase may introduce base mismatch, which leads to the difference between the synthesized cDNA sequence and the original RNA template. This will have a serious impact on some research that requires high sequence accuracy, such as gene editing and accurate gene expression analysis. In addition, the amplification efficiency of reverse transcriptase needs to be improved. For some RNA templates with low abundance, reverse transcriptase may not be able to convert them into cDNA efficiently, thus affecting the subsequent analysis and detection.
Biosafety risk: The application of reverse transcription technology involves the operation of biological genetic information, which also brings certain biosafety risks. When using retrovirus vector in gene therapy, if the design and operation of the vector are improper, it may lead to the accidental spread and spread of the virus, which may pose a potential threat to human health and ecological environment. In addition, the application of reverse transcription technology in laboratory research also requires strict safety management measures to prevent the disclosure and abuse of genetic information.
Unknown regulation mechanism: Although the basic process of reverse transcription has been understood to some extent, the regulation mechanism of reverse transcription is still poorly understood. In cells, the reverse transcription process is regulated by many factors, including protein factor and non-coding RNA. However, how these regulatory factors work together to accurately control the time, place and efficiency of reverse transcription is still an unsolved mystery. In-depth study on the regulation mechanism of reverse transcription is of great significance for understanding biological processes such as virus infection, cell differentiation and tumorigenesis.
Prospect of Reverse Transcription Research
Development of new reverse transcriptase: Through protein engineering technology, it is expected to develop a new reverse transcriptase with higher fidelity, amplification efficiency and thermal stability. These new reverse transcriptases will be able to synthesize cDNA more accurately and improve the reliability and sensitivity of reverse transcription technology. By reforming the structure of reverse transcriptase and optimizing its active center and substrate binding site, its performance may be significantly improved.
Innovation of detection technology: With the continuous development of sequencing technology and imaging technology, reverse transcription detection technology will also usher in innovation. Single cell sequencing technology can accurately detect RNA transcripts in a single cell, which is helpful to reveal the mechanism of cell heterogeneity and cell differentiation. High-resolution imaging technology can observe the dynamic changes of molecules in the process of reverse transcription in real time, which provides more direct evidence for in-depth study of the mechanism of reverse transcription.
Clarify the relationship between reverse transcription and diseases: Further study on the relationship between reverse transcription and diseases will provide new targets and strategies for the diagnosis and treatment of diseases. By studying the mechanism of reverse transcription in tumor occurrence and development, new anticancer drugs targeting reverse transcription related molecules can be developed. In addition, understanding the interaction between reverse transcription and immune system is also helpful to develop new immunotherapy methods and improve the therapeutic effect of diseases.
Replication fork progression and obstacles (Gómez-González et al., 2019)
Conclusion
The research of reverse transcription has made remarkable progress in many fields, such as the basic theory of biology, the application of biotechnology and medical clinical practice, but it also faces many challenges. From the in-depth analysis of the molecular mechanism of reverse transcription to the wide application of reverse transcription technology in genetic engineering, disease diagnosis and treatment, the development of this field has profoundly changed our ability to recognize and intervene in life processes.
For more related content, you may refer to the following articles:
- Overview of cDNA Synthesis
- cDNA: Current Applications and Future Horizons
- cDNA vs. gDNA: Structure, Function, Biotech Apps, and Why Choose cDNA
- Exploring cDNA: Introduction, Synthesis, gDNA Comparison, and Overcoming Challenges
- cDNA Library is Worth Your Trust
References
- Mougel M, Houzet L, Darlix JL. "When is it time for reverse transcription to start and go?" Retrovirology. 2009 6:24 https://doi.org/10.1186/1742-4690-6-24
- Hu WS, Hughes SH. "HIV-1 reverse transcription." Cold Spring Harb Perspect Med. 2012 2(10):a006882 https://doi.org/10.1101/cshperspect.a006882
- Zúñiga S, Sola I., et al. "Role of RNA chaperones in virus replication." Virus Res. 2009 139(2):253-66 https://doi.org/10.1016/j.virusres.2008.06.015
- Coté ML, Roth MJ. "Murine leukemia virus reverse transcriptase: structural comparison with HIV-1 reverse transcriptase." Virus Res. 2008 134(1-2):186-202 https://doi.org/10.1016/j.virusres.2008.01.001
- Menéndez-Arias L, Sebastián-Martín A, Álvarez M. "Viral reverse transcriptases." Virus Res. 2017 234:153-176 https://doi.org/10.1016/j.virusres.2016.12.019
- Gómez-González B, Aguilera A. "Transcription-mediated replication hindrance: a major driver of genome instability." Genes Dev. 2019 33(15-16):1008-1026 https://doi.org/10.1101/gad.324517.119


 Sample Submission Guidelines
Sample Submission Guidelines