Plant RNA Extraction Overview
In the extensive domain of molecular biology, the extraction of RNA from plants is recognized as a fundamental procedure in the study of gene expression, functional genomics, and transcriptomics. RNA,a conduit for genetic information, plays a pivotal role in various aspects of plant growth, development, and response to environmental stresses. However, the extraction of RNA from plants poses significant challenges, particularly given the complexity and diversity of plant tissues. The presence of high concentrations of secondary metabolites, polysaccharides, and phenolic compounds in plant tissues has been shown to interfere with the RNA extraction process, often resulting in RNA degradation or contamination. This, in turn, has been demonstrated to affect the results of subsequent experiments. Consequently, it is imperative to engage in a thorough discussion of the significance of plant RNA extraction, the challenges it faces, and effective extraction strategies, to facilitate the advancement of plant science research.
Commonly Used Plant RNA Extraction Methods
In the face of the intricacies inherent in the extraction of plant RNA, researchers have devised a range of methodologies, each with its distinct scope of application and advantages.
Phenol-Chloroform Method
The phenol-chloroform method remains a gold standard for RNA isolation, particularly in tissues rich in secondary metabolites. By leveraging the differential solubility of RNA, DNA, and proteins in phenolic solvents, this approach enables effective phase separation during centrifugation. However, the method is labor-intensive and demands stringent control of experimental conditions to prevent RNA degradation.
Ji et al. developed a protocol for RNA sequencing of PFA-fixed tissues, employing the phenol-chloroform method. Their approach integrated RNA extraction with the construction of random primer libraries, resulting in a substantial enhancement of RNA yield and quality. A comparative analysis revealed no significant disparities in sequencing quality metrics or detected gene profiles between fixed-tissue samples and matched fresh tissues. This method was successfully employed for region-specific gene expression analysis in the mouse brain.
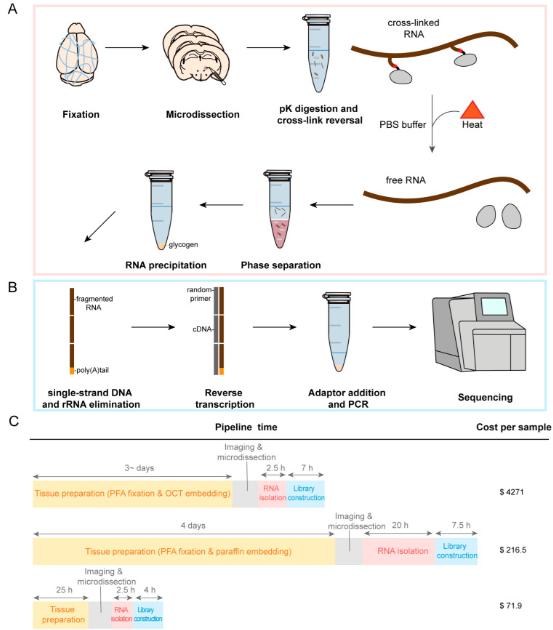
Sectioning and microdissection of regions of interest from PFA-fixed brain tissue; RNA is recovered using heat cross-link reversal with proteinase K in PBS buffer, followed by phenol-chloroform extraction and precipitation of total RNA with ethanol in the presence of sodium acetate and glycogen (Ji et al., 2024)
Trizol Method
The Trizol method represents a significant advancement in the field of RNA extraction. Notably, the Trizol method is characterized by its simplicity, rapidity, and broad applicability to diverse plant tissue types. The method is effective in obtaining high-quality RNA due to its ability to disrupt cell membranes, separate RNA from proteins and other cellular components, and through subsequent precipitation and washing steps. However, it should be noted that the Trizol reagent is relatively expensive and contains phenol, which is a known contaminant of the environment.
The Trizol platform was utilized by Choi et al. in the development of an innovative small RNA enrichment method, which they have termed "mirRICH". The validation of this method involved the use of sequencing and quantitative PCR, which confirmed its efficiency in the recovery of high-quality small RNAs from overdried precipitates. This advancement is significant in that it addresses critical challenges in the isolation of intact small RNAs from compromised samples.
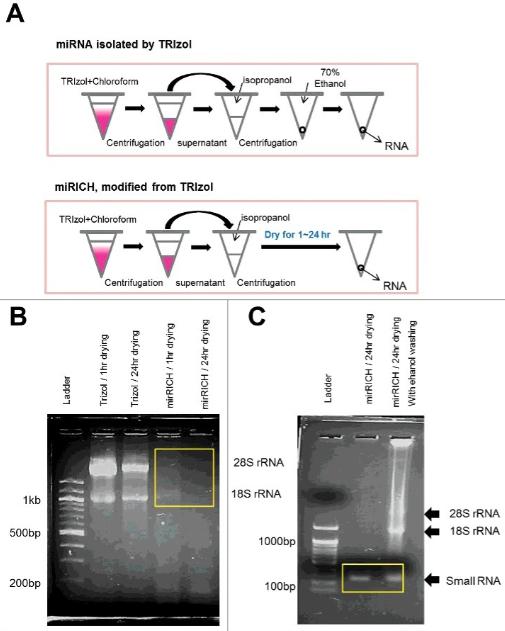
Over-drying based small RNA extraction method (Choi et al., 2024)
CTAB Method
The CTAB (hexadecyltrimethylammonium bromide) method has been specifically designed for the effective removal of polysaccharides and phenolic compounds from plant tissues. By forming insoluble complexes with these compounds, CTAB ensures the effective removal of contaminants while promoting the release and purification of RNA. Despite its efficacy, the procedure is technically demanding, requiring extended centrifugation and precipitation steps.
Badai et al. optimized this workflow by combining CTAB extraction with silica-based purification. Their protocol reduced extraction time from 48 hours to 5 hours. Quantitative RT-qPCR analysis demonstrated that total RNA isolated from 27 different oil palm tissues exhibited high integrity (RNA Integrity Number > 7), supporting its application in tissue-specific transcriptomic studies.
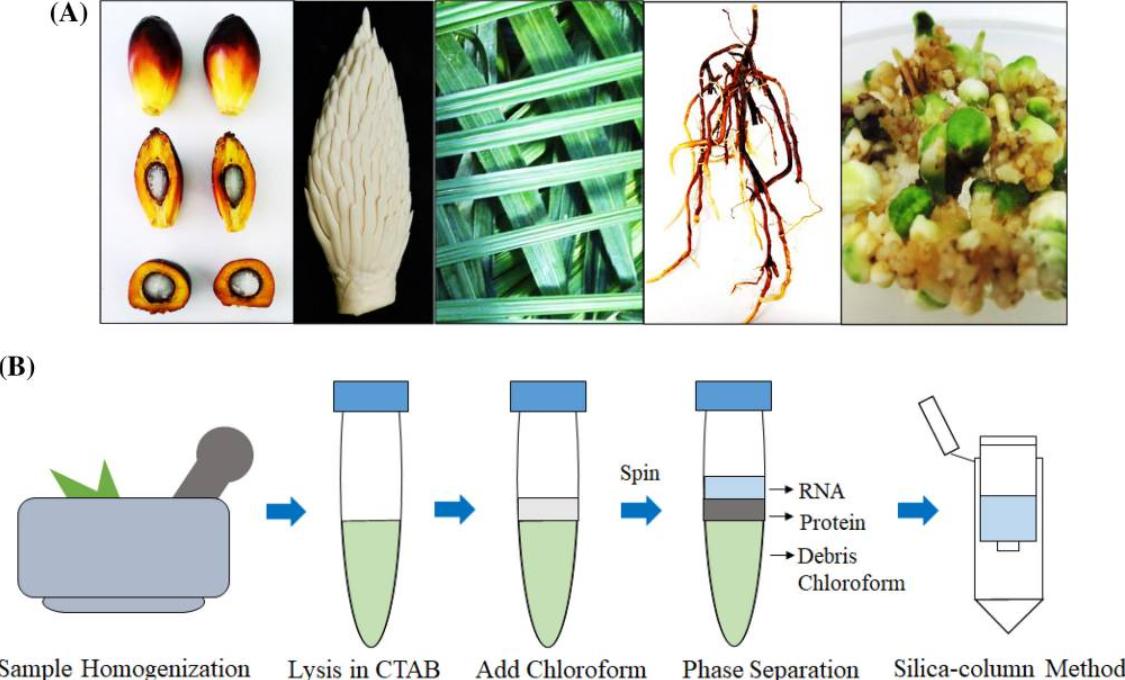
Overview of the total RNA extraction protocol using a combination of CTAB-based method and silica column (Badai et al., 2020)
Key Steps in Plant RNA Extraction
Successful RNA extraction from plants relies not only on the selection of an appropriate method but also on strict adherence to standardized operational procedures. The following are the critical steps involved in optimizing plant RNA extraction:
Sample collection and preservation
Freshly collected plant tissues should be processed or snap-frozen immediately to prevent RNA degradation. In the context of sample collection, it is imperative to exercise caution concerning instruments or containers that may contain RNase, to mitigate the risk of RNA degradation. For samples that are not to be processed immediately, it is essential to implement a rapid freezing process, followed by storage at minus 80 degrees celsius in a refrigerated environment. This approach is instrumental in preserving the integrity of the RNA.
Tissue homogenization
Tissue must be ground to a fine powder in liquid nitrogen, a critical step to ensure adequate cell fragmentation. The low temperature of liquid nitrogen rapidly freezes the water inside the cells, preventing RNase activity, and also aids in the mechanical fragmentation of the cells. The ground fine powder should be transferred immediately to a centrifuge tube containing the appropriate buffer to prevent the degradation of RNA.
RNA extraction and purification
The core step in RNA extraction and purification is the removal of proteins, DNA, and other impurities using appropriate buffers and reagents. Depending on the extraction method chosen, the addition of reagents such as phenol, chloroform, Trizol, or CTAB may be required to disrupt cell membranes, isolate RNA, and remove impurities. During the extraction process, temperature and time must be strictly controlled to avoid RNA degradation.
Precipitation and solubilization of RNA
Precipitation of RNA by isopropanol or ethanol represents a pivotal step in the process of obtaining high-quality RNA. Following this, the precipitated RNA should be solubilized using RNase-free ddH2O and subsequently purified and subjected to rigorous quality testing. During the dissolution process, vigorous vortexing or oscillation should be avoided to prevent RNA breakage.
Techniques to Enhance RNA Yield and Quality
Despite the established maturity of current RNA extraction methodologies, challenges persist regarding factors such as diminished RNA yield and compromised quality in practical applications. To address these challenges, it is imperative to enhance our comprehension and refine our competencies in RNA extraction.
Causes of RNA degradation
RNA degradation has been identified as a key factor affecting RNA yield and quality. The causes of its degradation mainly include contamination of RNase, high temperature, prolonged exposure to air, and inappropriate operation steps. It is notable that biological samples, including plant tissues and cells, contain significant levels of endogenous RNase, which is released following cell death or destruction, leading to RNA degradation. Additionally, exogenous RNase, present in dust, skin, hair, saliva, plastic gloves, and untreated experimental vessels, can also contribute to RNA degradation within the experimental environment. Furthermore, elevated temperatures have been shown to accelerate the RNA degradation reaction, resulting in RNA strand breakage. Consequently, the extraction process must be conducted under low temperatures. The ribose moiety of RNA molecules contains a hydroxyl group at the 2′ position, rendering them more vulnerable to attack by oxygen and water molecules in the ambient atmosphere. This susceptibility to oxidation and hydrolysis reactions is a significant consideration in biological research. Inappropriate experimental conditions, such as extended grinding times, elevated homogenization temperatures, and excessive centrifugation speeds, may also result in mechanical or chemical damage to RNA. To circumvent the deleterious effects of these unfavorable factors, it is imperative to meticulously regulate the experimental conditions.
Tips to improve RNA quality
To enhance the quality of RNA, it is essential to adopt the following techniques: firstly, it is imperative to utilize RNase-free instruments and containers during the procedure, and to refrain from any steps that may result in the introduction of RNase; secondly, it is crucial to maintain low-temperature conditions during the extraction process to reduce the activity of RNase. Secondly, the optimal extraction conditions are to be determined following the characteristics of the plant tissues and the extraction method selected, such as adjusting the pH value of the buffer, the ionic strength, the temperature, and the duration of the extraction process, to enhance the efficiency and purity of RNA extraction. Finally, the addition of RNA stabilizers, such as RNAase inhibitors and antioxidants, during the collection, processing, and extraction of samples, has been demonstrated to be an effective means of preventing RNA degradation and contamination.
RNA degradation represents one of the critical factors affecting the success of RNA-based experiments. A comprehensive understanding of its underlying mechanisms is essential for developing effective strategies to prevent it. By strictly controlling experimental conditions, using RNase-free instruments and containers, optimizing extraction parameters, and employing RNA stabilizers, we can significantly enhance RNA yield and quality. These technical improvements not only ensure the accuracy and reliability of downstream molecular biology assays but also provide a robust foundation for scientific research advancements.
Unlocking Plant RNA: Principles & Advances
General Principles
Successful RNA extraction from plants requires selecting appropriate methods based on tissue-specific characteristics and optimizing extraction conditions. When choosing an extraction protocol, comprehensive consideration should be given to factors including tissue composition, RNA quantity and quality requirements, simplicity of operation, and cost-effectiveness. By adhering to standardized procedures and incorporating proven techniques, high-quality RNA can be reliably obtained to serve as a solid foundation for subsequent experimental research.
Future Directions
Advancements in technology and the emergence of innovative methodologies will continue to enhance the efficiency and quality of plant RNA extraction. Emerging techniques such as magnetic bead-based methods and solid-phase extraction protocols have demonstrated significant potential in this field. These approaches offer advantages including streamlined workflows, rapid processing times, high-throughput automation, and reduced risk of contamination. Furthermore, as our understanding of plant RNA structure and function deepens, the demand for ultra-pure and highly intact RNA will grow. Consequently, the development of more efficient, sensitive, and specific extraction methods and reagents remains a critical focus for future research in plant molecular biology.
To further explore the methodologies and applications in this field, the following topics provide a comprehensive guide:
References
- Ji B, Chen J., et al. "Streamlined Full-Length Total RNA Sequencing of Paraformaldehyde-Fixed Brain Tissues" Int J Mol Sci. 2024 Jun 13;25(12):6504. https://doi.org/10.3390/ijms25126504
- Choi C, Yoon S., et al. "A simple method to enrich the small RNA fraction from over-dried RNA pellets." RNA Biol. 2018;15(6):763-772. https://doi.org/10.1080/15476286.2018.1451723
- Badai SS, Rasid OA., et al. "A rapid RNA extraction method from oil palm tissues suitable for reverse transcription quantitative real-time PCR (RT-qPCR)." 3 Biotech. 2020 Dec;10(12):530. https://doi.org/10.1007/s13205-020-02514-9


 Sample Submission Guidelines
Sample Submission Guidelines