Deciphering RNA-seq Library Preparation: From Principles to Protocol
Introduction
RNA sequencing (RNA-seq) has instigated a transformative shift in the sphere of molecular biology, facilitating scientists to explore gene expression profiles and regulatory mechanisms within cells with unparalleled precision. At the heart of RNA-seq's effectiveness lies the methodology of library preparation, a procedure crafted to transfigure RNA molecules into a form apt for high-throughput sequencing. In this review, we undertake an in-depth analysis of RNA-seq library preparation, meticulously dissecting its guiding principles, requisite stages, and the quantitative RNA thresholds for optimal library generation.
What is RNA-seq Library Preparation?
The initiation of RNA sequencing commences with RNA-seq library preparation, a pivotal and foundational phase within the RNA sequencing workflow. This crucial process orchestrates the transformation of RNA molecules into a library comprising DNA fragments amenable to interrogation via next-generation sequencing (NGS) platforms. The overarching objective of library preparation is multifaceted, aiming to safeguard the inherent biological information harbored within RNA molecules while concomitantly incorporating adaptors or barcodes essential for sequencing endeavors.
Commencing with the extraction of RNA from the pertinent biological specimen, the process advances through sequential stages. Initially, RNA molecules undergo fragmentation, resulting in the generation of smaller fragments conducive to downstream manipulations. These fragmented RNA molecules are subsequently transcribed in a reverse fashion into complementary DNA (cDNA) utilizing reverse transcriptase enzymes. The ensuing cDNA fragments are subjected to end repair, where any protruding ends are rectified, yielding DNA fragments with blunt termini. Following this, adaptors housing requisite sequencing motifs are ligated to the termini of the DNA fragments.
Ultimately, the constructed library undergoes enrichment to isolate fragments falling within the desired size range, accompanied by stringent quality control assessments to ensure fidelity and reliability prior to sequencing endeavors.
Detailed Protocol for RNA-seq Library Preparation
RNA sequencing library preparation stands as a pivotal juncture in the meticulous analysis of the transcriptome, necessitating scrupulous attention to detail to ensure the attainment of high-fidelity outcomes. In this exhaustive delineation, we offer a comprehensive protocol, delineating precise methodologies, requisite reagents, and pertinent considerations encapsulating each facet of the RNA-seq library preparation workflow.
RNA Isolation
Sample Collection: Employ aseptic techniques for the procurement of tissue or cell samples, promptly transferring them into RNase-free vessels to forestall RNA degradation.
Homogenization: Employ an appropriate method, such as tissue homogenization or bead milling, to disrupt cellular structures and facilitate RNA liberation.
RNA Extraction: Execute RNA extraction utilizing a commercial RNA isolation kit, adhering closely to the manufacturer's guidelines. Ensure robust DNase treatment to mitigate genomic DNA contamination.
RNA Quantification and Quality Assessment: Quantify the concentration and purity of the extracted RNA employing a spectrophotometer (e.g., NanoDrop) or fluorometer (e.g., Qubit). Evaluate RNA integrity through capillary electrophoresis (e.g., Bioanalyzer) or agarose gel electrophoresis.
RNA Fragmentation
Fragmentation Buffer Preparation: Prepare fragmentation buffer according to manufacturer specifications, tailoring fragmentation conditions to desired fragment sizes.
RNA Fragmentation: Introduce the isolated RNA into the fragmentation buffer, incubating under specified temperature and time parameters to achieve requisite fragment size distribution. Halt the reaction via addition of a stop solution or through thermal inactivation.
cDNA Synthesis
Reverse Transcription Reaction Setup: Assemble a master mix encompassing reverse transcriptase enzyme, random primers, dNTPs, and RNase inhibitor. Incubate fragmented RNA within the master mix at the appropriate temperature to facilitate cDNA synthesis.
cDNA Purification: Employ purification kits or magnetic beads to eliminate residual primers, enzymes, and salts from the synthesized cDNA. Evaluate cDNA yield and quality utilizing spectrophotometric or fluorometric methods.
End Repair and Adaptor Ligation
End Repair Reaction: Execute end repair by subjecting purified cDNA to end repair enzymes and buffers, monitoring reaction progression via gel or capillary electrophoresis.
Adaptor Ligation: Formulate a ligation mix comprising adaptors featuring unique barcodes or indices. Ligase adaptors to repaired cDNA termini under specified conditions. Subsequently, purify ligated products to eliminate unligated adaptors.
Size Selection and Enrichment
Library Fragment Size Selection: Employ gel electrophoresis, bead-based purification, or automated liquid handling systems for size selection of ligated products, isolating DNA fragments falling within the desired size range.
Library Amplification: Amplify size-selected library fragments via PCR, utilizing primers complementary to adaptor sequences. Optimize PCR conditions to mitigate amplification bias and ensure uniform coverage.
Library Quality Control
Library Quantification: Employ qPCR with adaptor-specific primers or fluorometric quantification to quantify amplified library.
Library Integrity Assessment: Evaluate size distribution and integrity of amplified library through capillary electrophoresis (e.g., Bioanalyzer) or agarose gel electrophoresis.
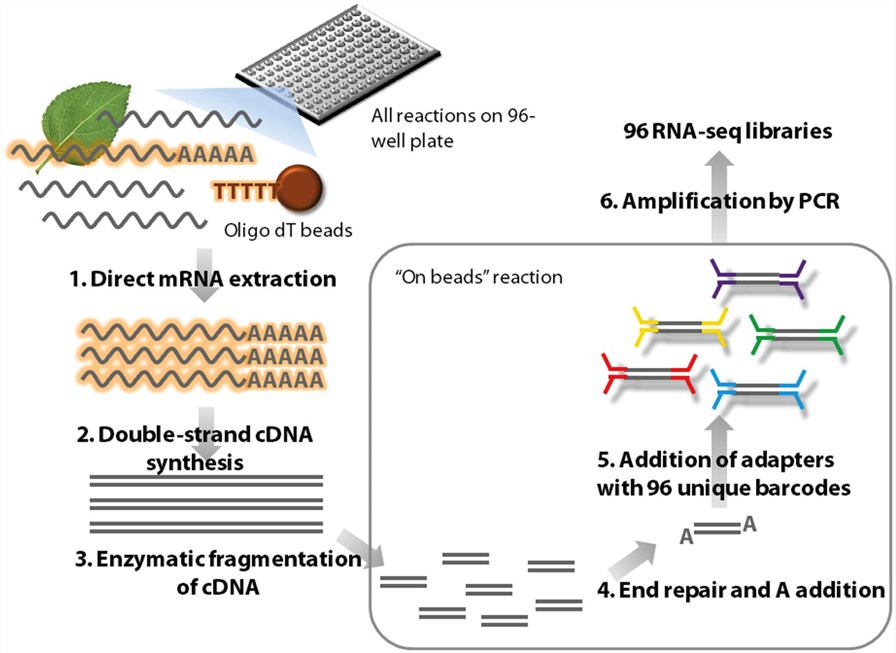 Outline of the high-throughput RNA-seq (HTR) library preparation.
Outline of the high-throughput RNA-seq (HTR) library preparation.
Amount of RNA Required for RNA-seq Library Preparation
The quantity of RNA requisite for RNA sequencing library preparation is contingent upon a multitude of variables, encompassing sample characteristics, RNA integrity metrics, sequencing platform specifications, and the desired depth of sequencing coverage. As a normative directive, a range of 0.1-1 μg of total RNA is advocated for the construction of high-fidelity RNA-seq libraries. Nonetheless, the optimal RNA input may exhibit variance contingent upon disparate experimental contexts and application domains.
For instances involving low-input RNA specimens or RNA of diminished integrity, specialized library preparation kits and protocols are procurable, tailored to surmount the impediments posed by restricted starting materials. These specialized kits harness sophisticated enzymatic modalities and amplification strategies to engender sequencing libraries from minute RNA quantities, all the while upholding library intricacy and curtailing inherent biases.
CD Genomics offers a comprehensive range of RNA-seq services, including library preparation, sequencing, and bioinformatics analysis, to support researchers in their quest to unravel the complexities of gene expression regulation. With our expertise and state-of-the-art technologies, we empower scientists to advance their understanding of biology and accelerate discoveries in various fields.
You may interested in
Learn More
Considerations for Optimizing RNA-seq Library Preparation
Refining RNA sequencing library preparation protocols stands as an imperative pursuit to uphold the fidelity and reproducibility of outcomes. A meticulous consideration of various factors is warranted during optimization endeavors, encompassing:
RNA Input Amount
The quantity of RNA input assumes paramount importance in RNA-seq library preparation. Inadequate RNA input may precipitate biased library construction and diminished sequencing depth, while excessive input may yield inefficient adaptor ligation and elevated background noise. Therefore, optimization of RNA input amounts necessitates a nuanced assessment, contingent upon sample attributes, RNA integrity metrics, and requisites of the chosen sequencing platform.
Fragmentation Method
The methodology employed for RNA fragmentation exerts notable influence on the size distribution and integrity of resultant cDNA fragments. Researchers are encouraged to scrutinize diverse fragmentation techniques, including sonication, enzymatic cleavage, or chemical fragmentation, to discern the most apt approach aligned with their experimental imperatives.
Adaptor Design
The architecture of sequencing adaptors holds sway over library complexity, sequencing coverage, and read quality. Customization of adaptor sequences featuring distinctive barcodes or indexes facilitates sample multiplexing and precise sample identification during subsequent data analysis. Careful selection of adaptor designs compatible with the designated sequencing platform and downstream bioinformatics workflows is advised.
Quality Control Measures
Imposing stringent quality control protocols across the entirety of the library preparation trajectory is indispensable to safeguard the integrity of sequencing data. Systematic evaluation of RNA integrity, cDNA fragment size distribution, and library concentration empowers the early detection of potential anomalies, thereby streamlining troubleshooting endeavors.
Bioinformatics Analysis
Proficient bioinformatics analysis serves as the linchpin for distilling meaningful insights from RNA-seq datasets. Rigorous data preprocessing, alignment to reference genomes or transcriptomes, and differential expression analyses constitute pivotal facets of the bioinformatics pipeline. Collaborative engagement with bioinformatics specialists or leveraging bioinformatics services offered by reputable entities such as CD Genomics can expedite the process of data analysis and interpretation.
FAQ of RNA-seq Library Preparation
Q: How much RNA is required for RNA-seq library preparation on the Illumina platform?
A: The amount of RNA required for RNA-seq library preparation on the Illumina platform depends on several factors, including the input material, sample type, and desired sequencing depth. However, a general guideline for total RNA input is typically in the range of 100 nanograms to 1 microgram.
Q: How to prepare RNA-seq data?
A: The RNA-seq protocol initiates with the conversion of RNA—be it total RNA, mRNA-enriched, or rRNA-depleted—into complementary DNA (cDNA). Following fragmentation, adapter ligation, and index ligation, each resultant cDNA fragment undergoes sequencing or "reading" utilizing a high-throughput platform.
Q: What is Illumina library preparation?
A: The preparation of Illumina libraries stands as a pivotal stage within NGS workflows, notably in the realm of RNA-seq investigations. This intricate procedure entails the transformation of RNA molecules into cDNA libraries amenable for sequencing on Illumina platforms. Typically encompassing RNA fragmentation, reverse transcription, adaptor ligation, and library amplification, Illumina library preparation protocols are meticulously crafted to yield libraries of exceptional quality characterized by uniform coverage and minimal bias. Such precision ensures the facilitation of precise and exhaustive transcriptome analyses.
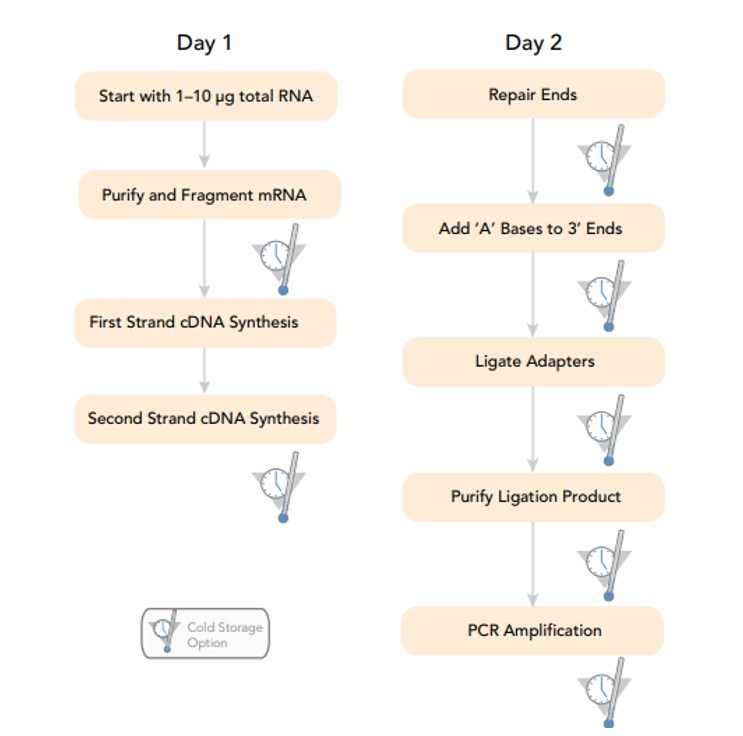 Illumina RNA-Seq Sample Preparation Workflow (from Illumina)
Illumina RNA-Seq Sample Preparation Workflow (from Illumina)
Q: How long does RNA-seq library preparation take?
A: The duration of RNA-seq library preparation exhibits variability contingent upon the chosen protocol, the nature of the sample, and the exigencies of the experiment. Typically, the process spans from several hours to a maximum of two days for completion. Standard library preparation methodologies entail a series of sequential enzymatic reactions encompassing RNA fragmentation, cDNA synthesis, adaptor ligation, and PCR amplification. The cumulative timeframe for library preparation encompasses hands-on benchwork, requisite incubation intervals, and optional quality control measures. Advanced automated systems for library preparation may expedite turnaround times by optimizing workflow efficiency and mitigating manual intervention.
Q: What is library size in RNA-seq?
A: In the context of RNA-seq experiments, the term "library size" pertains to the average length of cDNA fragments encompassed within the sequencing library. Within the ambit of library preparation, RNA molecules undergo fragmentation, yielding shorter segments, subsequent to which adaptors are ligated to both ends, thereby facilitating sequencing. The resultant library's size distribution assumes paramount significance for the successful execution of sequencing endeavors and the precision of subsequent data analyses. Evaluation of library size conventionally entails the utilization of capillary electrophoresis techniques (e.g., Bioanalyzer) or gel electrophoresis, furnishing insights into the breadth of fragment sizes encapsulated within the library. Optimal library size, contingent upon the sequencing platform and specific application, typically falls within the span of 200 to 500 base pairs for Illumina sequencing protocols.
Q: What is a library prep kit?
A: A library preparation kit, often denoted as a library prep kit, constitutes an exhaustive ensemble of reagents and protocols meticulously tailored to expedite the fabrication of sequencing libraries essential for NGS endeavors. These kits encompass a gamut of indispensable components, including enzymes, buffers, adaptors, and primers, pivotal for tasks spanning RNA or DNA fragmentation, cDNA synthesis, adaptor ligation, and library amplification.
Notably, CD Genomics offers a diverse array of RNA library preparation kits expressly designed for Illumina sequencing platforms. These kits are adeptly engineered to accommodate an array of sequencing applications and are calibrated to ensure optimal performance and reliability across diverse experimental paradigms. Researchers stand to gain from the flexibility inherent in selecting a library prep kit from CD Genomics that aligns seamlessly with their specific requirements, incorporating considerations such as sample type, input material characteristics, desired library intricacy, and compatibility with their preferred sequencing platform. The judicious selection of a CD Genomics library prep kit assumes pivotal significance in orchestrating the creation of high-fidelity sequencing libraries, thereby underpinning the acquisition of robust and dependable NGS data.
References:
- Kumar R, Ichihashi Y, Kimura S, Chitwood DH, Headland LR, Peng J, Maloof JN, Sinha NR. A High-Throughput Method for Illumina RNA-Seq Library Preparation. Front Plant Sci. 2012
- Ura, H., Togi, S. & Niida, Y. A comparison of mRNA sequencing (RNA-Seq) library preparation methods for transcriptome analysis. BMC Genomics 23, 303 (2022)


 Sample Submission Guidelines
Sample Submission Guidelines
