In the field of modern biology science, the continuous innovation of molecular biology technology promotes the exploration of gene function and regulation mechanism. As a synthetic DNA sequence, complementary DNA (cDNA) originated from the reverse transcription process of messenger RNA (mRNA), and has played an indispensable core role in many frontier research fields such as gene cloning, gene expression analysis and functional genomics since its birth.
This paper introduces cDNA in an all-round way, including its basic concept, differences from DNA, synthesis process, advantages compared with RNA, and analyzes the difficulties and limitations in the research. Furthermore, transcriptome sequencing (RNA-Seq), a key application of cDNA, is discussed as a powerful tool for profiling gene expression across different physiological or pathological conditions, offering dynamic insights into transcript-level regulation.
Introduction to cDNA
cDNA is a kind of single-stranded DNA which is complementary to mRNA and synthesized by reverse transcriptase. Then, through the action of DNA polymerase, it is transformed into double-stranded DNA, which is called cDNA. Eukaryotic genes usually contain introns and exons, while mRNA will remove introns and only retain exon sequences during post-transcriptional processing. Therefore, the cDNA synthesized by using mRNA as template does not contain introns, and its structure is more compact, which is convenient for operation and expression in genetic engineering and other fields. There is usually a poly A tail at the 3′ end of eukaryotic mRNA, and this structure will also be copied to cDNA during reverse transcription. Poly A tail has certain significance for the stability of cDNA and the subsequent experimental operation.
cDNA sequencing is a technique to determine the base sequence of cDNA obtained by reverse transcription. First, total RNA was extracted from biological cells, and then cDNA was synthesized by reverse transcriptase using mRNA as a template. Then, the cDNA was sequenced by high-throughput sequencing technique, and a large number of short sequence reads were obtained. After bioinformatics analysis, these reads can get the complete sequence information of genes, which can be used to study the structure, function, expression level and discover new genes, and have a wide range of applications in biology and medicine.
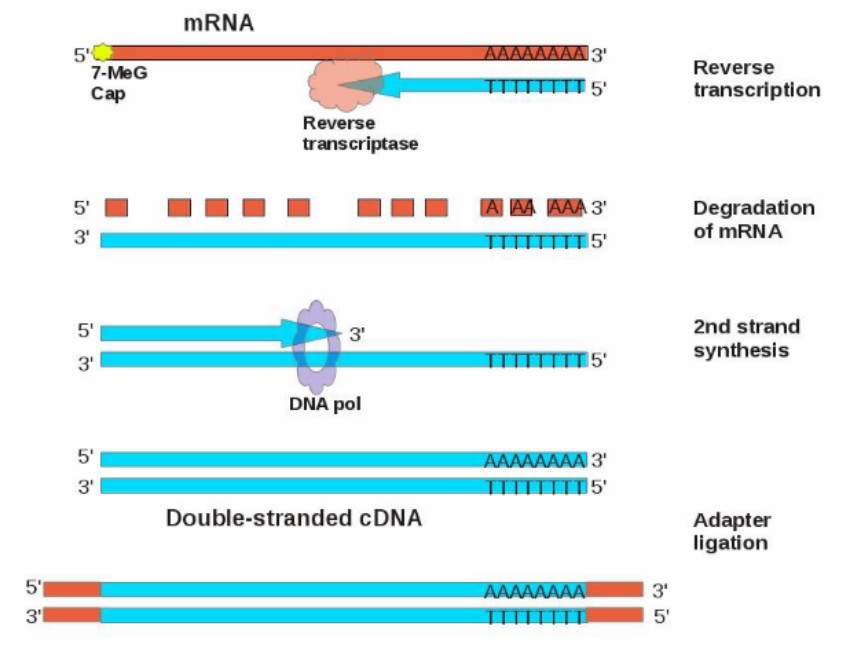
cDNA is formed via the synthesis from mRNA
How is cDNA Different from DNA
In the field of molecular biology, cDNA and DNA are two important concepts. Although they both carry genetic information, there are obvious differences in many aspects. Understanding these differences is crucial for further exploring gene function, disease mechanism and developing related biotechnology.
DNA, namely deoxyribonucleic acid, is the main genetic material of an organism and exists in the nucleus (eukaryote) or the eukaryote (prokaryote). It consists of two anti-parallel deoxynucleotide chains, which form a double helix structure through the principle of base complementary pairing. The nucleotide sequence of DNA contains all the genetic instructions needed to construct and maintain the life activities of organisms, and its length varies from species to species. The human genome DNA has about 3 billion base pairs. DNA can not only stably transmit genetic information to daughter cells through semiconservative replication, but also be transcribed into RNA, thus guiding the synthesis of protein.
cDNA does not exist naturally in cells, but is artificially synthesized in the laboratory through reverse transcription. Reverse transcription takes mRNA as a template, and single-stranded DNA complementary to mRNA is synthesized by reverse transcriptase, and then double-stranded cDNA is synthesized by DNA polymerase. This means that the synthesis of cDNA depends on the existence of mRNA. Because mRNA is the product of DNA transcription, and after splicing, the intron sequence in the gene is removed, so cDNA only contains the coding region (exon) of the gene, without intron.
Structurally, DNA is usually a double-stranded linear or circular structure, while cDNA is generally a double-stranded linear structure, and its length is shorter than that of the corresponding gDNA, because it lacks non-coding sequences such as introns. In terms of function, DNA bears the function of storing and transmitting genetic information, and is the core substance of organism heredity. cDNA is mainly used in the field of biotechnology. When studying gene expression, cDNA can reflect the genes actually expressed in specific cells or tissues under specific physiological conditions, while DNA contains all genes in cells, whether they are expressed or not.
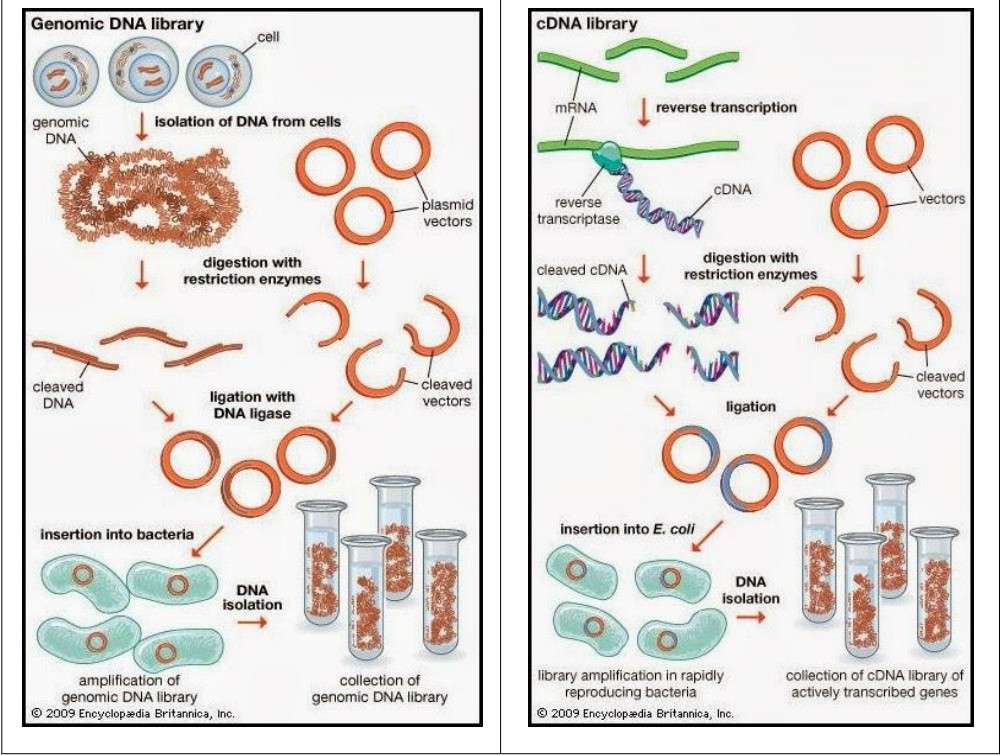
Different cDNA and gDNA library construction
The Process of cDNA Synthesis
cDNA synthesis is the process of reverse transcription of mRNA into cDNA by reverse transcriptase.
Fundamental Principle
Using mRNA as a template, its poly-A can be used as a primer binding site. Adding a segment of oligo-dT as a primer, which can combine with the poly-A tail of mRNA complementarily, and under the action of reverse transcriptase, using four kinds of deoxynucleoside triphosphates (dNTP) as raw materials, according to the principle of base complementary pairing, a DNA chain complementary to mRNA is synthesized from the 3′-OH end of the primer, forming an RNA-DNA hybrid double strand. Then the RNA chain is degraded by alkali treatment or RNase H, and then the remaining DNA chain is used as a template to synthesize the second DNA chain under the action of DNA polymerase, and finally the double-stranded cDNA is obtained.
key Steps
In the research of molecular biology, In-depth study and understanding of the key steps involved in cDNA synthesis is better to understand this core technology.
RNA extraction: Total RNA is extracted from cells or tissues, and then mRNA is purified by means of oligocellulose column chromatography.
Primer design and addition: According to the characteristics of mRNA, design and add appropriate primers, such as oligo-dT primers or specific primers for specific genes.
Reverse transcription reaction: Mix mRNA, primer, reverse transcriptase, dNTP, etc. in an appropriate buffer, incubate at a certain temperature, perform reverse transcription reaction, and generate the first strand of cDNA.
Second strand synthesis: The second strand of cDNA was synthesized by different methods, such as using DNA polymerase I and RNase H, etc.
Purification and detection of cDNA: Purify cDNA by column chromatography and gel electrophoresis, and then detect the quality and yield of cDNA.
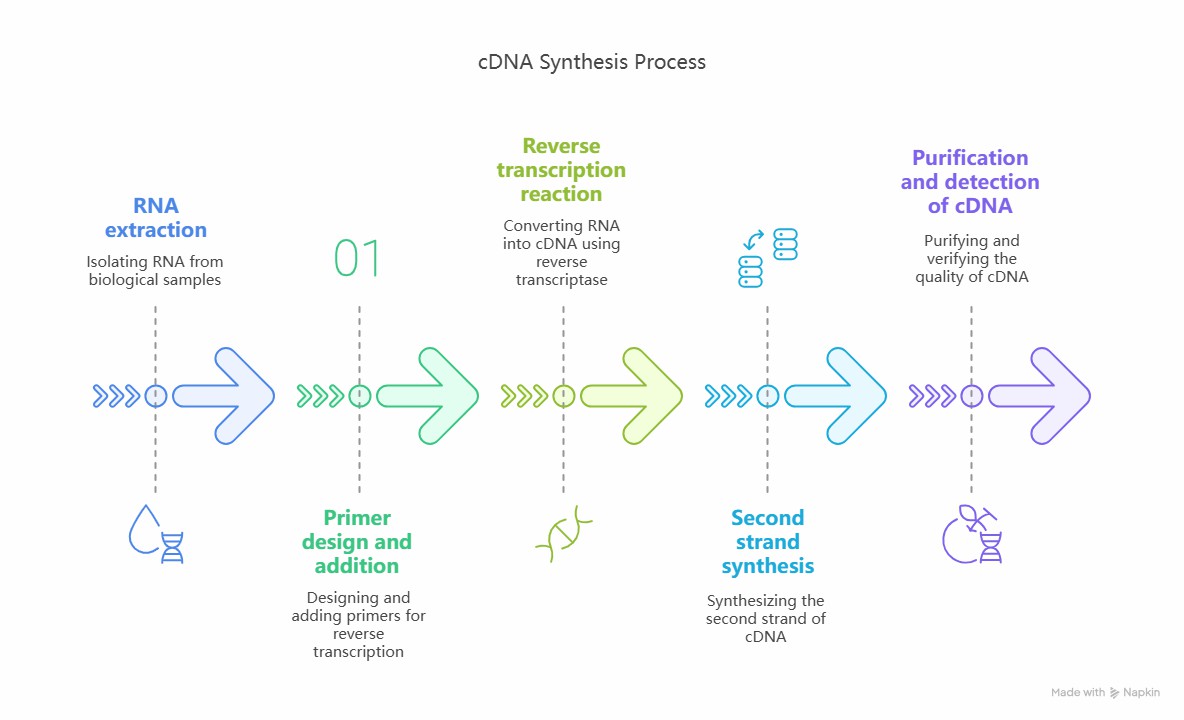
Workflow of cDNA synthesis
Why is cDNA used instead of RNA
In the field of molecular biology, researchers often choose to use cDNA instead of RNA to carry out research work. This selection is based on the remarkable advantages of cDNA compared with RNA in many aspects, such as stability, convenience of operation, and being used for cloning and amplification.
Stability Aspect
RNA is a relatively unstable molecule. It has a single-stranded structure, which makes it more vulnerable to nuclease attack. Nucleases exist widely in the environment and cells, and can degrade RNA molecules rapidly. In the laboratory environment, even if strict anti-enzyme measures are taken, RNA may be degraded in a short time, thus affecting the accuracy of experimental results.
In sharp contrast, cDNA is a double-stranded DNA molecule. The double-stranded structure endows cDNA with higher chemical stability, which enables it to be stored and manipulated under relatively loose conditions. At the temperature of -20℃ or even higher, cDNA can remain stable for a long time and is not easily degraded by nuclease. This makes it easier for researchers to store, transport and post-process the cDNA during the experiment, which greatly improves the repeatability and reliability of the experiment.
Operational Convenience
Manipulation of RNA requires very strict experimental conditions. Because RNA is easily degraded, it is necessary to use RNase-free consumables and reagents in the process of extraction, purification and analysis, and to operate in a special experimental environment to prevent RNase pollution. This undoubtedly increases the difficulty and cost of the experiment.
The operation of cDNA is relatively simple. Once RNA is converted into cDNA by reverse transcription reaction, the subsequent operations can be carried out according to the conventional DNA experiment method. In the molecular biology experiments such as PCR amplification, restriction enzyme digestion and ligation, the operation of cDNA is basically the same as that of ordinary DNA, and no special anti-enzyme measures are needed. This enables researchers to complete the experiment more efficiently and reduces the errors and uncertainties in the experiment process.
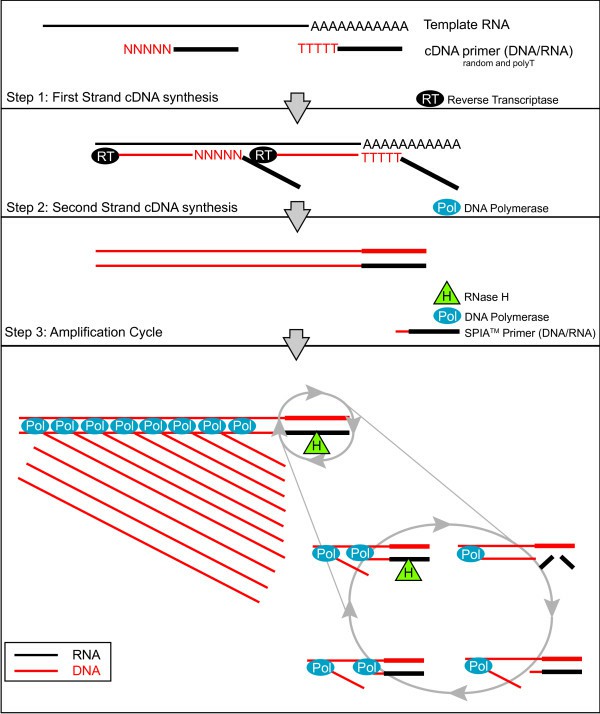
Diagram of the ribo-SPIA process according to cDNA synthesis (Watson et al., 2008)
Gene Cloning and Amplification
RNA cannot be directly used for cloning and amplification. Because most cloning and amplification techniques, such as PCR and molecular cloning, are designed based on the double-stranded structure of DNA and the principle of base complementary pairing. The single-stranded structure and chemical properties of RNA make it impossible to be directly applied to these technologies.
cDNA can be easily used for cloning and amplification. Through PCR technology, a large number of specific cDNA fragments can be amplified, so as to obtain enough samples for subsequent analysis. In the molecular cloning experiment, cDNA can be inserted into a suitable vector and introduced into a host cell for expression and research. This enables researchers to deeply understand the function and regulation mechanism of genes.
Gene Expression
In gene expression analysis, although RNA can directly reflect the transcription level of genes, it is difficult to accurately measure the expression level of RNA because of its instability and complexity of operation.
cDNA provides a more reliable tool for gene expression analysis. After RNA is transformed into cDNA by reverse transcription, the expression level of specific genes can be accurately measured by real-time quantitative PCR (qPCR) and other technologies. In addition, cDNA can also be used to construct a gene expression library. By sequencing and analyzing the cDNA in the library, we can fully understand the gene expression in cells or tissues. This method can more accurately reflect the gene expression changes under different physiological and pathological conditions, and provide an important basis for the diagnosis, treatment and drug development of diseases.
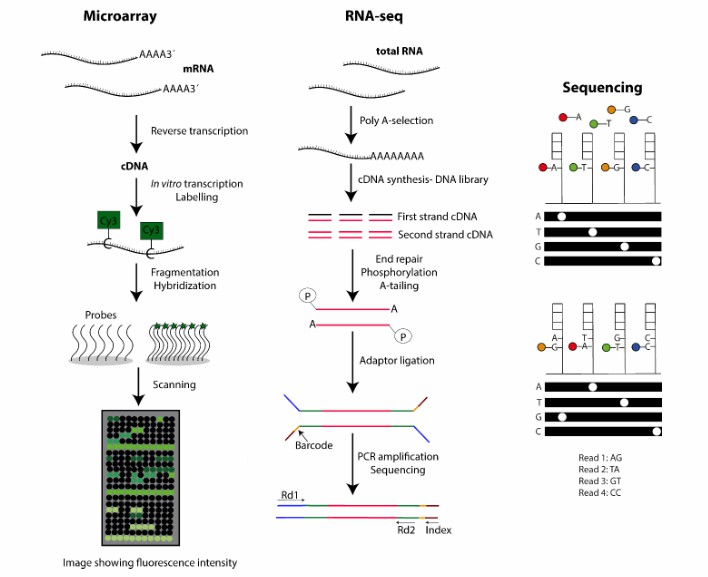
Comparison of sequencing method based on cDNA and RNA
Challenges and Limitations in Working with cDNA
cDNA technology provides a key research method for the exploration of gene function, the analysis of disease mechanism and the research and development of biopharmaceuticals. It represents the transcription activity of cells in a specific period. However, like any powerful technical tool, in the process of using cDNA to carry out research, researchers also face a series of thorny challenges and obvious limitations.
Difficulties in cDNA Synthesis
CDNA synthesis is the initial key step of the whole research process, but it is easy to encounter many obstacles. Firstly, the integrity of mRNA has a decisive influence on the quality of cDNA synthesis. In the process of cell lysis and subsequent extraction, mRNA molecules are very fragile and easily degraded by endogenous RNase. Even a tiny amount of RNase pollution may lead to the break of mRNA chain, resulting in the deletion of fragments in the synthesized cDNA, which can not fully reflect the original transcript information.
The selection and performance of reverse transcriptase are also very important. Different reverse transcriptases differ in activity, affinity to templates and extensibility. When some reverse transcriptases encounter mRNA regions rich in secondary structure, they are easy to stop or terminate the reaction in advance, which greatly reduces the synthesis efficiency of long-chain cDNA. Taking some mRNA with high GC content as an example, its complex secondary structure often discourages reverse transcriptase, and it is difficult to successfully synthesize a complete corresponding cDNA.
In addition, primer design is also an easily overlooked but far-reaching factor. Although random primers can widely bind to mRNA and start the reverse transcription reaction, it may lead to short synthetic cDNA fragments and high sequence randomness. Although specific primers can accurately target the target mRNA, they need to know its sequence accurately. Once the primer design is unreasonable, such as mismatch and dimer formation, it will seriously affect the specificity and efficiency of cDNA synthesis.
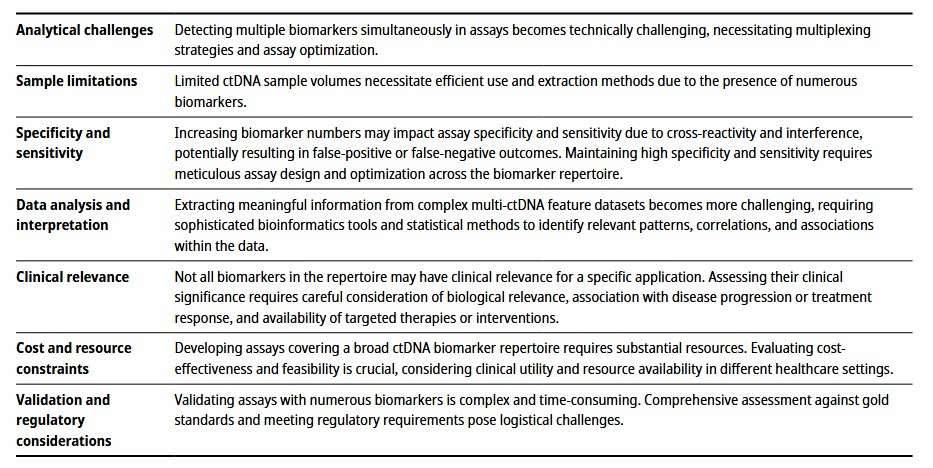
Difficulties of cDNA-based assays (Bronkhorst et al., 2023)
The Challenge of cDNA Library Construction
Constructing high-quality cDNA library is an important basis for in-depth study of gene expression profile and function, but this process is equally difficult. In the process of library construction, it is a big challenge to ensure the complete preservation of cDNA diversity. Due to the huge difference of mRNA abundance in cells, high-abundance mRNA is easy to dominate in the process of library construction, which makes the number of cDNA copies corresponding to low-abundance mRNA extremely low or even difficult to detect.
Linking efficiency is another key issue in library construction. Connecting the synthesized cDNA fragment to the vector is not a smooth process. The connection between cDNA fragment and the end of vector requires accurate restriction site matching and efficient ligase catalysis. In practice, the efficiency of ligation is often restricted by many factors, such as improper molar ratio of cDNA to vector, interference of impurities in ligation reaction system, etc., which will lead to ligation failure or wrong ligation, resulting in a large number of empty vectors or clones with abnormal recombinant structure in the library, which greatly increases the workload of subsequent screening and identification.
Limitations in cDNA Analysis
There are also many limitations in the follow-up analysis of cDNA. From the quantitative point of view, quantitative methods based on cDNA, such as qPCR, are highly sensitive, but their accuracy is easily affected by many factors. The difference of cDNA synthesis efficiency between different samples and the inconsistency of primer amplification efficiency may lead to large deviation of quantitative results.
In functional research, although cDNA represents the coding region of genes, it lacks peripheral information such as gene regulatory elements. It is impossible to fully understand the true function of genes in the complex regulatory network in vivo by studying cDNA only. In addition, with the deepening of research, the demand for single-cell cDNA analysis is increasing, but the single-cell cDNA research is facing more severe challenges. The mRNA content in a single cell is extremely low, so it is easy to introduce amplification deviation in the process of cDNA amplification, which leads to the wrong amplification or reduction of gene expression differences between different cells, and it is difficult to accurately describe the transcriptome map of a single cell.
Conclusion
cDNA synthesis, as a key means to obtain complementary DNA from mRNA, has laid the foundation for further research. With the catalysis of reverse transcriptase, cDNA synthesis skillfully bypasses the complex structure of genomic DNA introns and accurately captures gene expression information, which opens a door for exploring the molecular mechanism of life activities.
RNA sequencing (RNA-seq) technology provides an important solution to overcome the limitations of cDNA analysis, and RNA-seq is especially suitable for the detection of low-abundance transcripts. Because high-abundance mRNA tends to dominate in the process of cDNA synthesis, the information of low-abundance transcripts is lost, while RNA-seq can cover transcripts with different abundance more evenly through deep sequencing.
For more related content, you may refer to the following articles:
- Overview of cDNA Synthesis
- cDNA: Current Applications and Future Horizons
- cDNA vs. gDNA: Structure, Function, Biotech Apps, and Why Choose cDNA
- Reverse Transcription: An In-Depth Exploration of Structure, Function and Application
- cDNA Library is Worth Your Trust
References
- Nygaard, Vigdis and Eivind Hovig. "Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling." Nucleic Acids Research 34 (2006): 996-1014 https://doi.org/10.1093/nar%2Fgkj499
- Chaudhuri, Anusha et al. "Rrp6p/Rrp47p constitutes an independent nuclear turnover system of mature small non-coding RNAs in Saccharomyces cerevisiae." bioRxiv (2020): 13 https://doi.org/10.1101/2020.12.13.422512
- Watson JD, Wang S., et al. "Complementary RNA amplification methods enhance microarray identification of transcripts expressed in the C. elegans nervous system." BMC Genomics. 2008 19 9:84 https://doi.org/10.1186/1471-2164-9-84
- Ozsolak F, Milos PM. "RNA sequencing: advances, challenges and opportunities." Nat Rev Genet. 2011 12(2):87-98 https://doi.org/10.1038/nrg2934
- Bronkhorst AJ, Holdenrieder S. "The changing face of circulating tumor DNA (ctDNA) profiling: Factors that shape the landscape of methodologies, technologies, and commercialization." Med Genet. 2023 35(4):201-235 https://doi.org/10.1515/medgen-2023-2065
- Semenkovich NP, Szymanski JJ., et al. "Genomic approaches to cancer and minimal residual disease detection using circulating tumor DNA." J Immunother Cancer. 2023 11(6):e006284 https://doi.org/10.1136/jitc-2022-006284


 Sample Submission Guidelines
Sample Submission Guidelines