In the field of molecular biology, complementary DNA (cDNA) and genomic DNA (gDNA) are two crucial concepts, which play a key role in life science research, biotechnology application and many other aspects. Although both of them are related to the transmission and interpretation of genetic information, there are significant differences in structure, function and acquisition methods. A deep understanding of these differences is of far-reaching significance to accurately grasp the molecular mechanism of life process and carry out efficient biotechnology research and development.
This paper compares the differences between cDNA and gDNA in structure, function and biotechnology application, and expounds the advantages of cDNA in avoiding intron interference, reflecting gene expression status and efficiently expressing specific protein. In this context, transcriptome sequencing (RNA-Seq) is widely used to analyze cDNA and explore dynamic gene expression patterns, while genome sequencing provides comprehensive insights into the full genetic blueprint encoded in gDNA.
What is cDNA and gDNA
gDNA refers to the complete genomic DNA in the cell of an organism, which contains all the genetic information of the organism. It exists in the nucleus in the form of chromosomes (eukaryotes) or directly in cells (prokaryotes). Human gDNA is distributed on 23 pairs of chromosomes, containing about 3 billion base pairs, encoding tens of thousands of genes and a large number of non-coding sequences with regulatory functions. From the evolutionary point of view, gDNA carries the genetic code accumulated by long-term evolution of species, which determines the basic characteristics and genetic traits of organisms.
cDNA is a DNA molecule synthesized by reverse transcription with mRNA as a template. In eukaryotes, when genes are expressed, DNA is transcribed to generate mRNA, which leaves the nucleus and enters the cytoplasm after a series of processing, and serves as a template for protein synthesis in the cytoplasm. Researchers use reverse transcriptase, using mRNA as template, and synthesize complementary DNA chain, namely cDNA, according to the principle of base complementary pairing. This process reverses the flow direction of genetic information from DNA to RNA, so it is called reverse transcription. For example, when studying the genes actively expressed by a specific cell in a certain physiological state, the mRNA of the cell can be extracted, and then the cDNA can be synthesized for subsequent analysis.
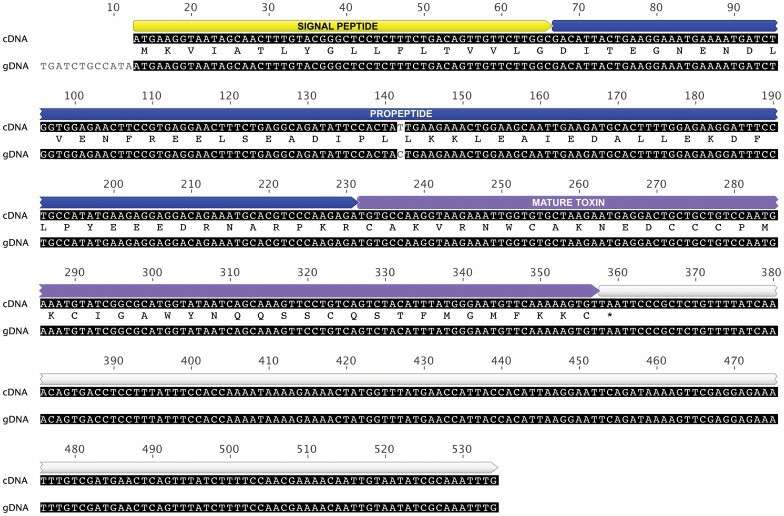
Alignment of the cDNA and gDNA sequences obtained for d-hexatoxin-Hi1a (Pineda et al., 2012)
Structural Differences Between cDNA and gDNA
There are significant differences between gDNA and cDNA. gDNA represents complete genetic information of organisms, including complex structures such as exons and introns. cDNA, on the other hand, is reverse transcribed from mRNA, and usually only contains exon sequences encoding protein.
The Existence of Exons and Introns
The structure of gDNA is complex, which contains all the components of genes, including exons and introns. Exons are regions of genes that encode protein, while introns are non-coding sequences located between exons. In the process of gene transcription, DNA is first transcribed into precursor mRNA, which contains information of exons and introns. Then, the precursor mRNA is spliced to remove introns and connect exons to form mature mRNA.
cDNA only contains exon sequences. Because it is reverse transcribed from mature mRNA, and mature mRNA has removed introns. This makes the structure of cDNA relatively simple, and only contains the information needed to encode protein. When studying eukaryotic genes, there may be multiple introns in gDNA sequence to separate exons, while the corresponding cDNA sequence is a continuous exon sequence.
Gene Regulation
gDNA contains not only the coding region, but also a large number of gene regulatory elements, such as promoters, enhancers, silencers and so on. Promoter is a DNA sequence located upstream of the gene, which is the binding site of RNA polymerase and can start the transcription process of the gene. Enhancers and silencers can play a role far away from the gene, and enhance or inhibit the expression level of the gene by interacting with transcription factors. These regulatory elements are very important for the precise regulation of gene expression, and they can regulate the transcription activity of genes according to the physiological state, development stage and external environmental signals of cells.
cDNA usually does not contain these gene regulatory elements. Because it is reverse transcribed from mRNA, the information of regulatory elements is not preserved during the post-transcriptional processing of mRNA. Therefore, cDNA is mainly used to study the coding sequence and protein expression of genes, while gDNA is usually used to study the regulation of gene expression.
Repetitive Sequence
There are a large number of repetitive sequences in gDNA, which can be divided into different types, such as satellite DNA, microsatellite DNA and scattered repetitive sequences. Satellite DNA usually consists of short repeating units ranging in length from hundreds to millions of base pairs, which are mainly distributed in the centromere and telomere regions of chromosomes. Microsatellite DNA consists of shorter repeating units (usually 1-6 base pairs) and is widely distributed in the genome. Scattered repetitive sequences are sequences that can move in different positions in the genome, such as transposons. These repetitive sequences play an important role in the structure, function and evolution of genome, but they also increase the complexity of gDNA structure.
cDNA generally does not contain repetitive sequences, because mRNA mainly encodes protein, and repetitive sequences usually do not participate in protein coding. Therefore, the sequence of cDNA is relatively pure, which is more suitable for gene cloning and expression analysis.
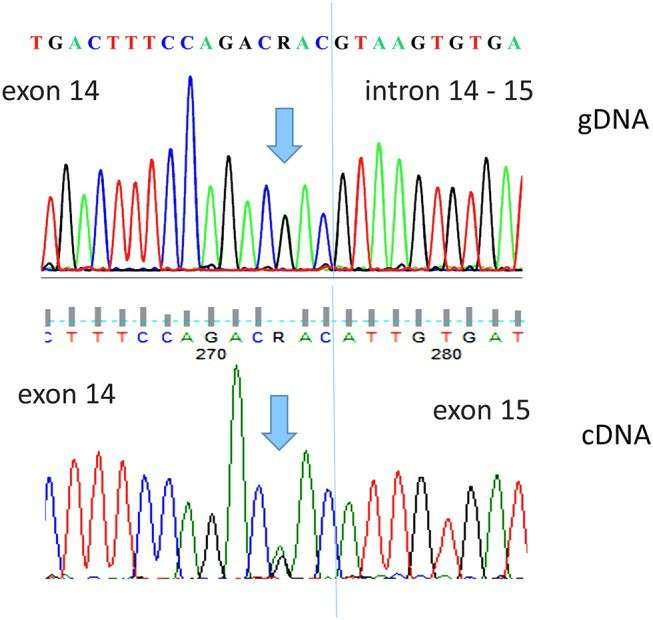
Direct sequence analysis of the DPYD c.1903A>G, p.Asn635Asp variant (Palmirotta et al., 2019)
Different Function and Role in the Cell
In the microscopic world of cells, although both cDNA and gDNA are related to genetic information, they play completely different roles and perform unique functions in cells.
The Role of gDNA in Cells
Storage of genetic information: gDNA is a complete repository of genetic information in cells. It contains all the instructions needed for all life activities such as growth, development and reproduction of organisms. Taking human cells as an example, gDNA is distributed on 23 pairs of chromosomes, with about 3 billion base pairs. The arrangement and combination of these base pairs constitute tens of thousands of genes and a large number of non-coding sequences with regulatory functions.
Transmission of genetic information: gDNA plays a key role in the transmission of genetic information during cell division. Through the mechanism of DNA replication, an accurate copy of gDNA of the parent cell is transmitted to the daughter cell, which ensures that the daughter cell inherits the genetic characteristics of the parent cell. This transmission of genetic information ensures the genetic stability and continuity of species and enables organisms to maintain their basic biological characteristics in generations.
The regulatory center of gene expression: gDNA contains not only the coding sequence of genes, but also many regulatory elements, such as promoters, enhancers and silencers. Promoter is located in the upstream of gene, which is the site of RNA polymerase recognition and binding, and it determines the start of gene transcription. Enhancers and silencers can enhance or inhibit gene expression by interacting with specific transcription factors at positions far away from genes. These regulatory elements work together to accurately regulate the expression levels of different genes in cells at different times and in different tissues, so that cells can adapt to various internal and external environmental changes.
The Role of cDNA in Cells
Reflect the gene expression state: cDNA is reverse transcribed from mRNA, which represents the gene information actually expressed by cells at a specific moment. Unlike gDNA, cDNA only contains the coding region (exon) of the gene, without intron. Therefore, by analyzing cDNA, researchers can know which genes are actively expressed in specific cells or tissues and their expression levels.
Template Mediator of protein Synthesis: In the cell, the synthesis of protein needs mRNA as a template. However, cDNA and mRNA are complementary. In the field of biotechnology, cDNA can be used as an important tool to realize the in vitro synthesis of protein. The researchers cloned the cDNA into an appropriate expression vector, and then introduced it into a host cell (such as E.coli, yeast or mammalian cells), so that the host cell can synthesize the corresponding protein according to the information carried by the cDNA. This method has been widely used in the field of biopharmaceuticals.
A powerful tool for studying gene function: cDNA library is an important resource for studying gene function. By constructing a cDNA library of a specific cell or tissue, researchers can screen out the genes of interest and study their functions in depth. Through the experiment of gene deletion or overexpression in cDNA library, we can understand the specific role of these genes in the process of cell growth, differentiation and metabolism.
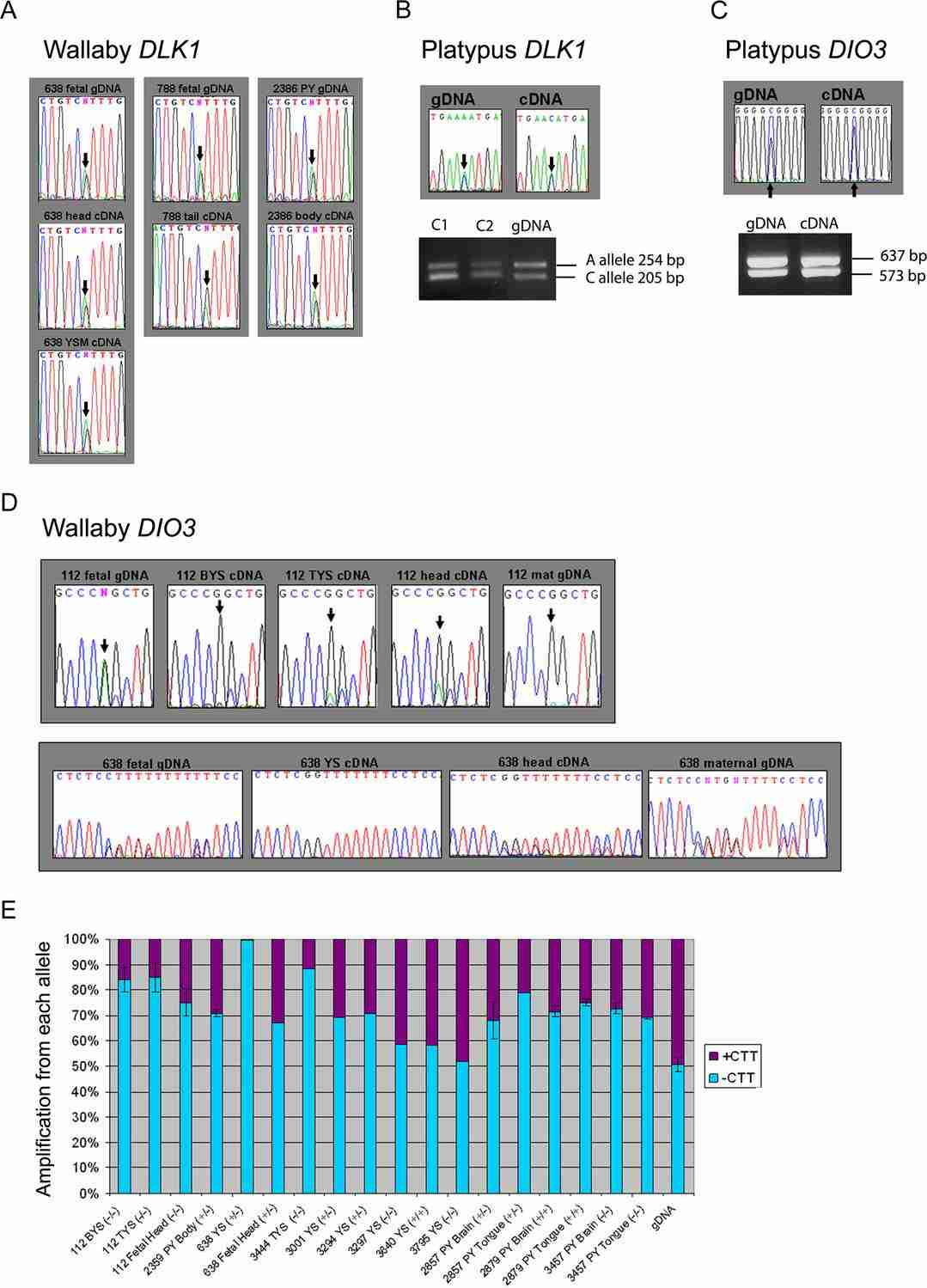
Biallelic Expression of DLK1 and DIO3 in the form of cDNA and gDNA (Edwards et al., 2008)
Cooperate to Maintain the Normal Function of Cells
Although there are obvious differences in function and function between cDNA and gDNA, they cooperate with each other to maintain the normal physiological function of cells. As the source of genetic information, gDNA provides stable genetic information for cells. cDNA, on the other hand, is the product of gDNA’s expression in a specific environment. According to the needs of cells, it transforms the genetic information in gDNA into actual protein and performs various biological functions.
cDNA and gDNA have different functions and roles in cells, and their synergy ensures the normal growth, development and functional maintenance of cells. The in-depth study of cDNA and gDNA not only helps us to reveal the mystery of life, but also provides an important theoretical basis and technical means for the development of biotechnology and medicine.
Applications of cDNA and gDNA in Biotechnology
As the key elements of biotechnology research, cDNA and gDNA have different characteristics and sources, which determines their unique advantages and limitations in various biotechnology applications. The comparison of the application of cDNA and gDNA in biotechnology is helpful for researchers to make better use of these two genetic materials and promote the development of biotechnology in medicine, agriculture, industry and other fields.
Gene Expression Analysis
cDNA is a commonly used material in gene expression analysis. The expression level of specific genes can be accurately measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR). cDNA library can be used for large-scale gene expression profile analysis to identify differentially expressed genes.
Although gDNA itself cannot directly reflect the gene expression, it can be used to study the regulatory elements of genes. By sequencing and analyzing gDNA, we can identify regulatory sequences such as promoters and enhancers, and understand how they affect gene expression. In addition, gDNA methylation analysis is also an important means to study gene expression regulation, and the change of methylation status can affect the transcription activity of genes.
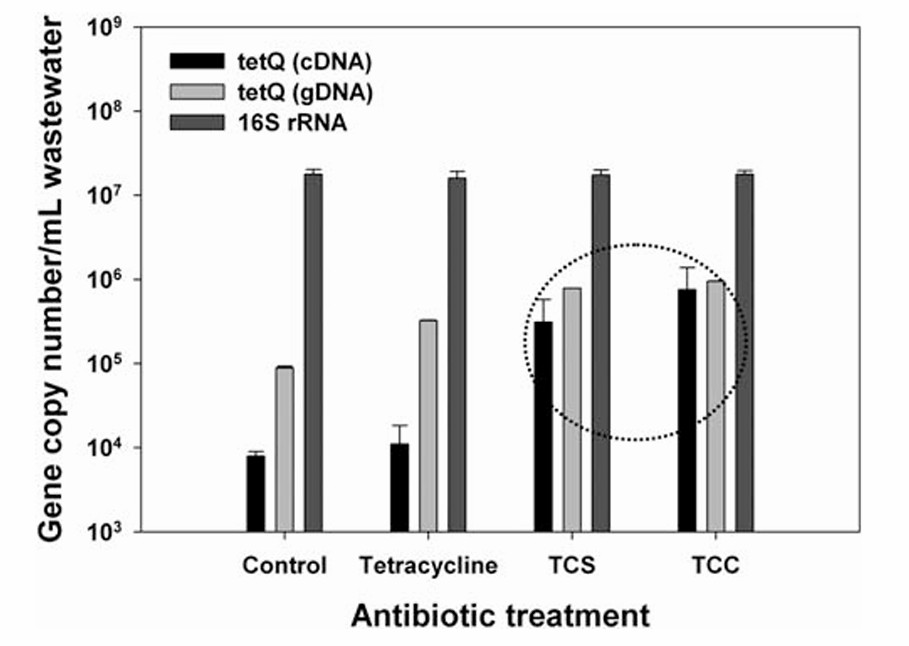
Comparison between cDNA gene expression and gDNA quantification using MLNPs-DNA assay (Son et al., 2010)
Gene Therapy
In gene therapy, cDNA is often used to replace defective genes or introduce new therapeutic genes. Because the cDNA sequence is relatively small, it is easier to be packaged into virus vectors to achieve efficient gene delivery. In the treatment of some monogenic diseases, normal cDNA can be introduced into patients’ cells to correct gene defects. In addition, cDNA can also be used to construct RNA interference (RNAi) vectors to inhibit the expression of harmful genes.
The application of gDNA in gene therapy is relatively rare, but it has unique advantages in some cases. For some therapeutic schemes that require complete gene regulation, it may be more appropriate to use gDNA fragments containing regulatory sequences. In addition, gDNA editing technology (such as CRISPR-Cas9) can directly modify the genome, which provides a new way to treat some complex genetic diseases.
Application of Molecular Diagnosis
In molecular diagnosis, cDNA is often used to detect the change of expression level of specific genes to assist the diagnosis and prognosis evaluation of diseases. In the diagnosis of breast cancer, detecting the cDNA expression level of some genes related to tumor metastasis can predict the invasion of tumor and the prognosis of patients. In addition, cDNA can also be used to detect virus infection, and the existence and replication level of virus can be determined by detecting the cDNA obtained by reverse transcription of virus mRNA.
gDNA is mainly used to detect genetic variation, such as single nucleotide polymorphism (SNP), insertion deletion (InDel) and copy number variation (CNV). These genetic variations are closely related to the occurrence and development of many diseases. By sequencing and analyzing gDNA of patients, accurate diagnosis and personalized treatment of diseases can be realized.
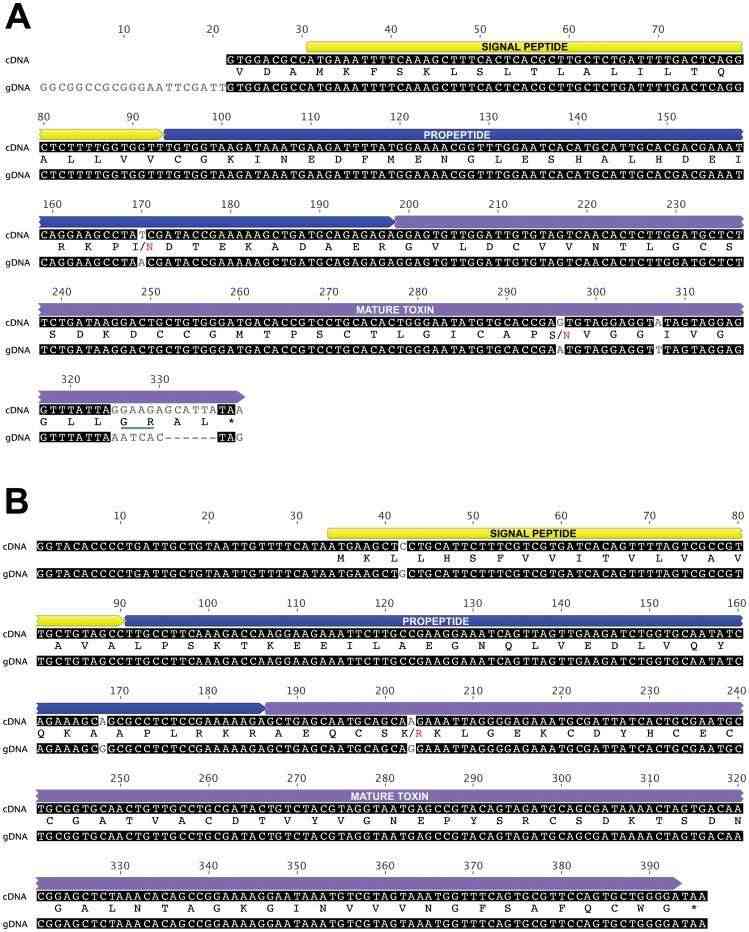
Architecture of cDNA and gDNA sequences of (A) v-hexatoxin-Hi2a and (B) U 3-hexatoxin-Hi1 (Pineda et al., 2012)
Why Choose cDNA for Certain Studies
In molecular biology, when it comes to certain types of research, cDNA often stands out and becomes the first choice tool for researchers. This is closely related to the unique structure, function and convenience in experimental operation of cDNA.
Avoid Intron Interference
As a complete representative of the genome, the coding region (exon) of gDNA is separated by many non-coding introns. However, in many research scenarios, the existence of introns is not necessary, but it becomes an obstacle in the research process. The birth of cDNA cleverly avoided this problem. It is synthesized by reverse transcription using mRNA as template. Because the intron has been removed and only the exon sequence has been retained in the post-transcriptional processing stage, mRNA is essentially a pure version of the gene coding region.
Accurately Reflect the Gene Expression Status
Under the stimulation of different physiological states, developmental stages and external environment, gene expression is constantly changing, and cDNA plays an irreplaceable role in reflecting gene expression, because it is a mirror copy of mRNA actively transcribed in cells, and each cDNA corresponds to a gene being expressed. Through the analysis of cDNA, researchers can intuitively and accurately understand the gene expression profile of cells at a specific moment. In the field of tumor research, by comparing the cDNA libraries of normal cells and tumor cells, we can clearly find the genes with abnormally high or low expression in tumor cells.
Efficient Expression of Specific Proteins
It is of great economic value and social significance to efficiently express specific protein in vitro by means of genetic engineering, which is used in drug research and development, vaccine production, industrial enzyme preparation and other fields. cDNA plays a central role in this process. Because it has no intron, compact structure and only contains the key sequence encoding protein, after cloning it into an appropriate expression vector and introducing it into a host cell (such as Escherichia coli, yeast or mammalian cells), the host cell can transcribe and translate it more efficiently, thus avoiding the problems such as transcription abnormality and splicing errors that may occur when dealing with complex gDNA structures.
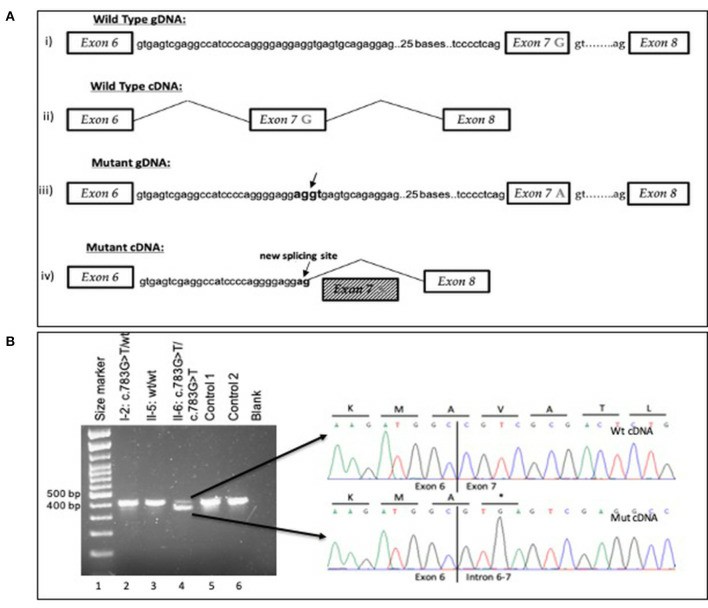
Schematic overview of the wild-type and mutant gDNA and cDNA of exons 6–8 in the ARPC1B gene (Papadatou et al., 2021)
In the study of cDNA, RNA sequencing (RNA-seq) technology can fully analyze the transcriptome characteristics of cells in a specific state by reverse transcription of mRNA into cDNA and then high-throughput sequencing. For gDNA research, genome-wide sequencing (WGS) technology can reveal the structural variation, single nucleotide polymorphism (SNP) and copy number variation (CNV) of the genome. These data are very important for understanding the molecular mechanism of genetic diseases and developing accurate diagnosis and treatment programs.
Conclusion
There are significant differences between cDNA and gDNA in source, structure, function, acquisition mode and application field. gDNA plays a cornerstone role in the inheritance of species genetic information, genetic analysis and species identification. cDNA, on the other hand, focuses on genes that are actively expressed in specific time and space, and shows unique advantages in gene function research and biotechnology product development.
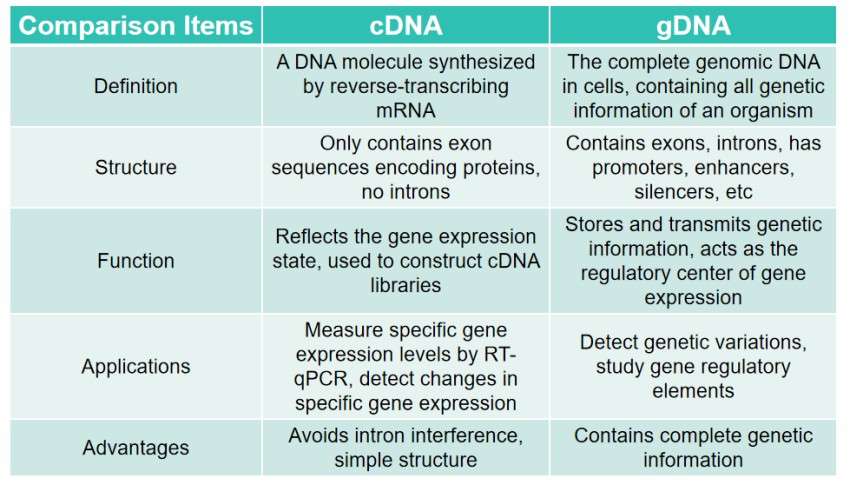
Comparison of gDNA and cDNA
For more related content, you may refer to the following articles:
- Overview of cDNA Synthesis
- cDNA: Current Applications and Future Horizons
- Exploring cDNA: Introduction, Synthesis, gDNA Comparison, and Overcoming Challenges
- Reverse Transcription: An In-Depth Exploration of Structure, Function and Application
- cDNA Library is Worth Your Trust
References
- Kubo M, Nishiyama T., et al. "Single-cell transcriptome analysis of Physcomitrella leaf cells during reprogramming using microcapillary manipulation." Nucleic Acids Res. 2019 47(9):4539-4553 https://doi.org/10.1093/nar/gkz181
- Palmirotta R, Lovero D., et al. "Rare Dihydropyrimidine Dehydrogenase Variants and Toxicity by Floropyrimidines: A Case Report." Front Oncol. 2019 9:139 https://doi.org/10.3389/fonc.2019.00139
- Edwards CA, Mungall AJ., et al. "The Evolution of the DLK1-DIO3 Imprinted Domain in Mammals." PLoS Biol 6(6): e135 https://doi.org/10.1371/journal.pbio.0060135
- Pineda SS, Wilson D., et al. "The lethal toxin from Australian funnel-web spiders is encoded by an intronless gene." PLoS One. 2012 7(8):e43699 https://doi.org/10.1371/journal.pone.0043699
- Son M, Kennedy IM., et al. "Quantitative gene monitoring of microbial tetracycline resistance using magnetic luminescent nanoparticles." J. Environ. Monit. 2010 12 1362-1367 https://doi.org/10.1039/C001974G
- Papadatou I, Marinakis N., et al. "Case Report: A Novel Synonymous ARPC1B Gene Mutation Causes a Syndrome of Combined Immunodeficiency, Asthma, and Allergy With Significant Intrafamilial Clinical Heterogeneity." Front Immunol. 2021 12:634313 https://doi.org/10.3389/fimmu.2021.634313


 Sample Submission Guidelines
Sample Submission Guidelines